New Contenders for Anticancer Therapeutics: Microbial Ribonucleases
Rakesh Kumar and Shamsher S Kanwar*
Department of Biotechnology, Himachal Pradesh University, India
Submission: September 20, 2017; Published: February 27, 2018
*Corresponding author: Shamsher S Kanwar, Department of Biotechnology, Himachal Pradesh University, India.
How to cite this article: Rakesh Kumar, Shamsher S Kanwar. New Contenders for Anticancer Therapeutics: Microbial Ribonucleases. Adv Biotech & Micro. 2018; 8(3): 555739. DOI: 10.19080/AIBM.2018.08.555739
Abstract
Microbial ribonucleases (RNases) are small molecules which are highly cytotoxic in nature. They catalyze the degradation of cellular RNA into oligonucleotides and mononucleotides. The cytotoxic properties of RNases include degradation of RNA leading to blockage of protein synthesis in malignant cells and induction of the apoptosis response. Cytotoxicity of RNases is determined by catalytic activity, stability, non-selective nature of inhibitors, positive charge on molecule and internalization. Binase, Barnase, RNase Sa, RNase P and other microbial RNases exert cytotoxic activity on cancer cells selectively by involving different cellular pathways and they also boost the cytotoxicity by chemical modification or mutation. Microbial RNases have general mechanism of cytotoxicity mediated by their interaction with the cellular membrane by nonspecific interactions mediated by Columbic forces, internalization by endocytosis, translocation to the cytosol, degradation of ribonucleic acid and leads to cell death by activation of caspase-dependent mechanisms, alteration or modification of target protein and NF-κB signal pathway. But still it is unclear that which of these suggested mechanisms cause cell death in malignant cells. This article looks at the mechanism of cytotoxicity of microbial RNases towards the cancerous cells which makes RNase to be considered as a prominent chemotherapeutic or anticancer agent.
Keywords: Microbial ribonuclease; Cytotoxicity; Cellular pathways; Apoptosis; Anticancer therapeutics
Introduction
All microorganisms contain different classes of RNases that are involved in RNA degradation. RNA degradation is indispensable step(s) in the cellular and biological processes. Cellular RNA molecules must endure the specific cleavage reactions in order to accomplish their mature functional forms by ribonucleases (RNases). Microbial RNases are hydrolytic enzymes that help in degradation of phosphodiester bond of cellular RNA. Studies on RNases began when Walter Jones discovered a thermostable enzyme in the extracts of bovine pancreas that hydrolyzed transforming yeast RNA into soluble acid product without discharging of inorganic phosphate or pyrimidine and purine bases [1,2]. In 1924, nucleic acid degrading enzyme was found in commercial Takadiastase sourced from Aspergillus oryzae [3]. Both RNase and DNase activity were observed in 36 strain of hemolytic group of Streptococci RNase activity was also observed in actinomyctes culture and the presence of RNA-degrading enzymes was also seen in many other fungi. Ribonuclease T1 was first microbial RNase isolated from commercial enzyme mixture from A. oryaze [4] and other fungal species. Their substrate specificity revealed that presence of two cyclic RNases from Takadiastase which splited specifically phosphodiester bond of 3’-gaunylic acid in RNA and smaller component have specificity for phosphodiester bonds of 3’-adenylic acid [4]. The primary structure of RNase T1 was determined and well characterized by X-ray crystallography, NMR spectroscopy and chemical modification. A guanalic acid-specific RNase was known as RNase T1, and an adenylic acid- specific RNase was named as RNase T2 [4]. RNases with varying specificities have been reported from microbes, plants and animals [2].
Microbial RNase have ultimate capability to degrade cellular RNA into oligonucleotides and mononucleotides besides presenting many other biological activities like maturation and decay reactions, which exhibit diverse structures and mechanisms of action (Figure 1). Microbial RNases are heterogeneous group of enzymes which may be grouped broadly into two classes according to the approach in which they cleave the cellular RNA. Endoribonucleases cleave specifically the cellular RNA internal phosphodiesters, while exoribonucleases cleave RNA chains from the 3P or the 5P terminus creating mononucleotide products (Figure1 ) [5-7].
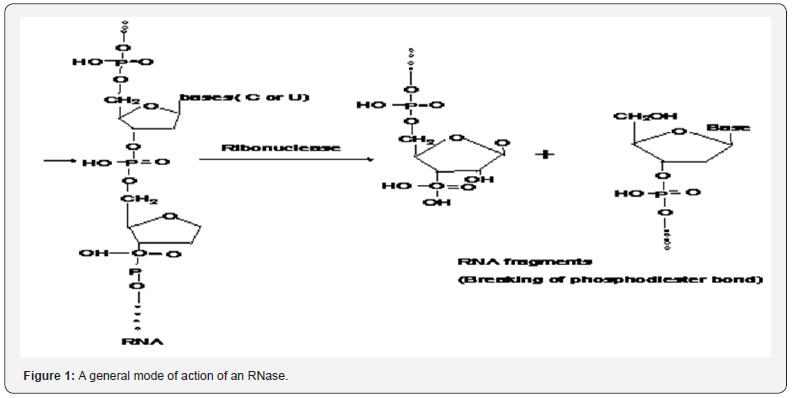
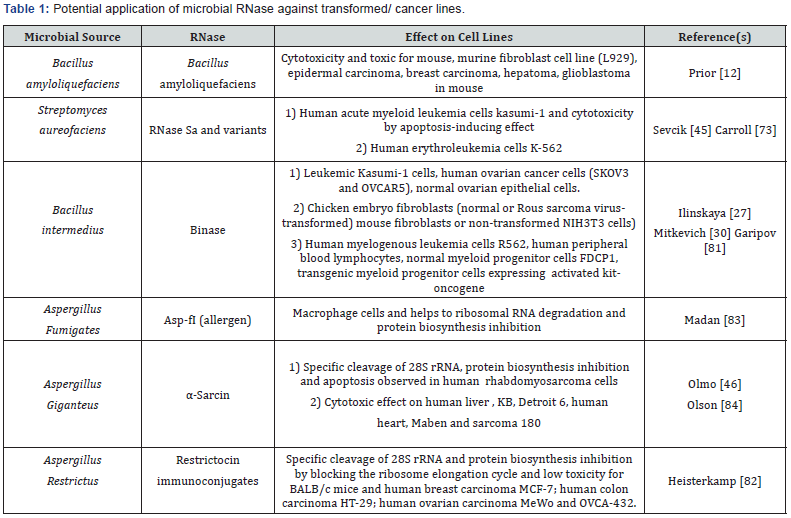
Exoribonucleases and endoribonucleases are further divided into several sub-classes. The major type of exoribonuceases are PNPase, RNase II and oligoribonuclease for RNA degradation others such as RNase PH, RNase BN, RNase D and RNase T are mainly responsible for the 3’end maturation of RNA. Major microbial endoribonucleases have been classified into RNase I, RNase III, RNase P, RNase E, RNase HI, HII and HIII [5,7,8]. Interestingly, often microbial RNases are highly cytotoxic towards in vitro cultured malignant cells. Quite often RNases trigger apoptotic response due to this property, and they are thus also being considered as alternative chemotherapeutic drugs (Table 1).
In case of animal or human RNases, the enzyme loses its catalytic activity due to chemical modification to some extent. Cationic variants of bovine RNase A and human RNase 1 generated by chemical modification of carboxyl groups exhibited no cytotoxicity if their catalytic activity was less than 0.01% of the non-modified enzyme [9]. Onconase ribonucleolytic activity is caspase-dependent apoptosis in target cells leads to damage of cytoplasmic t-RNA [10,11]. An RNase from Bacillus amyloliquefaciens known as ‘Barnase’ attached to the transport domain of Pseudomonas exotoxin-A for infiltration of malignant cells was observed to exert toxic effect(s) against several malignant cell lines in vitro [12]. Over a period of time the RNases have been found to be potential antitumor agents based on their existing cytotoxic activities in vitro and in vivo.
The idea of using RNases as antitumor agents came into sight in year 1955 when scientists found the RNA as key intermediate of protein synthesis of any cell [13]. By that time, initial experiments were performed by using pancreatic RNase because of its availability in purified and large amounts. The mammalian RNase demonstrated antitumor activity in vitro, in vivo and also in patients. However, outcomes of these experimental results could not be consistently reproduced, and researchers turned their attention to other sources of RNases. RNases from microbial sources seem promising as well. In year 1965, an RNase isolated from the Aspergillus giganteus was named as α-Sarcin. α-Sarcin was found to be active/ cytotoxic against model sarcomas in rats [14].
Unfortunately, clinical studies of microbial enzymes as well as some other RNases encountered serious difficulties due to low efficiency and toxicity and had to be discontinued. In 1980, the eurkaryotic RNase known as Onconase showed the antitumor activity and was observed to have cytotoxic and cytostatic effect [15]. Yet, final results of the Phase III trial were made public due to ethical issues and some drawbacks. In this article, we turn our attention to microbial RNases as an alternative biomolecules for novel anticancer therapy. Although the first paper demonstrating antitumor activity of bacterial nuclease from Serratia marcescens appeared in 1964, however the development of bacterial RNases into antitumor drugs for many years remained almost uncharted realm [16]. Meanwhile, the RNases are a large and diverse group of proteins unrelated to eukaryotic enzymes with a different cytotoxicity profile. In particular, among many bacterial RNases, the bacterial T1 RNases which demonstrated killing-activity against malignant cells was preferred as its RNase activity could be enhanced by genetic manipulations and chemical modifications.
Microbial ribonucleases as strong contenders as anticancer agents
Microbial RNases are a large and diverse group of proteins unrelated to eukaryotic enzymes and with a different cytotoxicity profile. Although, first article that demonstrated the antitumor activity of microbial RNase from Serratia marcescens came into prominence about six decades ago [16], however the development of microbial RNases into antitumor drugs until now remained almost an unattended option [17]. The following section will emphasize the use of microbial RNases as alternative and novel anticancer therapeutic agents.
RNase T1: RNase T1 is type of an endonuclease which degrades phosphodiester bonds of cellular RNA between 3’-guanylic residues and 5’-OH residues of adjoining nucleotides with the development of consequent intermediate 2’,3’- cyclic phosphates. RNase T1 has been iso¬lated mainly from Aspergillus and Penicillium species. RNase T1 has cytotoxic nature towards malignant cells without metal ions, and because of this character it is also known as Ribotoxin [18]. RNase T1 has been used to analyze RNA structure, mapping, RNA protection and removal of RNA form extracted DNA samples. RNase T1 from Aspergillus oryzae and RNases U1 from Ustilago sphaerogena were inactivated by RI [19,20]. RNase T1 shows toxic effects by targeting the conserved sequence known as Sarcin-ricin loop of larger rRNA and thus it leads to inhibition of protein biosynthe¬sis in the target cell followed by eventual cellular death by apoptosis. RNase T1 internalizes into cell-cytosol by Columbic interaction or Clathrin-independent protein. The Ribotoxin protein that targets cellular RNA eventually leads to cytotoxic effect towards the tumor cells. Ribotoxins preferentially kill the tumor cells by altered membrane permeability if no specific protein receptor has been present on the cell membrane, such as the cells infected with virus or transformed cells [15,18].
RNase T1 is consisting of a small protein α + β dimer containing 104 amino acid residues with Cys2-Cys10 and Cys6- Cys103 disulfide bonds help in folding and unfolding of RNase T1 [21]. RNase T1 specifically targets the guanine bases of cellular RNA for denaturing. Researchers have found that HVJ as vector can be targeted to tumour cells to make it to act as a unique anticancer drug. RNase T1 has been internalized by cells via a novel gene transfer reagent, hemagglutinating virus of Japan (HVJ) envelope vec¬tor into human tumor that resulted in the tumour cell death. Researchers observed that pre-treatment of HVJ envelope vector with protamine sulphate which leads RNase T1 showed tremendously improved cytotoxic activity. RNase T1 is in phase III human clinical trials as a non-mutagenic cancer chemotherapeutic agent [22]. Furthermore, internalized RNase T1 also induces the apoptotic cell death program in the tumour cells. However, previous studies have indicated that the RNase T1 cytotoxicity is unfortunately not specific to tumor cells but it could otherwise be overcome by HVJ envelope vector thus making the RNase a highly specific antitumor tool [23,24].
Binase: Binase, a low-molecular weight (12.2-kDa) protein containing 109 amino acids has been isolated from Bacillus intermedius. Binase is well studied in various aspects including its purification, sequencing, crystal structure to in vitro and in vivo antitumor activities [25,17]. Binase unit contains two molecules forming a homodimer, and one of the subunits can bind the nucleotide in the 3′GMP-Binase complex in which the guanyl base is located in the recognition loop of the enzyme [26]. Binase revealed a broad spectrum and persuasive activity against malignant cells. In previous studies, it has been reported that Binase has antiproliferative and selective apoptosis-inducing activities against human myelogenic erythroleukemia K562 cells, peripheral blood mononuclear cells and human lung carcinoma A549 cells [27]. However, Binase-mediated antiproliferative and apoptotic effects were not detected in normal human peripheral blood. In transformed myeloid cells, apoptosis was induced by Binase but it however, didn’t induce the T-cell immune response [28]. Binase attacked artificially expressing activated c-Kit myeloid progenitor follicular dendritic cells (FDC) and chronic myelogenous leukemia K562 cells, but did not induce apoptosis in normal myeloid progenitor cells and leukocytes of healthy donors [28].
Extracellular cytotoxic ribonucleases (RNases) of bacterial origin, that trigger apoptosis/cytotoxicity in tumor cells, may have a great potential as anticancer drugs [17,29,30]. A Binase sourced from Bacillus pumilus was selectively cytotoxic towards lung carcinoma, leukemic and ovarian cancer cells. Interestingly, it was found that the cell sensitivity towards xenogeneic RNases is related to expression of oncogenes ras, kit and AML1 [31]. Binase selective apoptotic properties found towards malignant cells confirmed that it could be considered as an alternative antitumor chemotherapeutic drug [32,33]. The Binase is completely unrelated to any eukaryotic RNases, and hence it is not susceptible to omnipresent eukaryotic RNase inhibitors (RI). Binase possesses remarkable biological activities which exert positive mutagenic properties at high concentrations and at low concentration it stimulates the cell growth. The most fascinating feature of Binase is its antitumor activity but the specific mechanism(s) of antitumor activity remains unknown. These encouraging observations unveil the therapeutic potentials of bacterial RNases, especially Binase for the treatment of tumors mononuclear cells.
Barnase: Barnase is also a promising contender found as an anticancer therapeutic molecule which possesses 84% sequence homology with Binase and is also the closest natural homologue of Binase. Barnase was initially isolated from Bacillus amiloliquefaciens as an active proenzyme. It was processed by the removal of the amino-terminal signal peptide, and was secreted into the extracellular space. It showed promising results against target cancer cells. Importantly, it had low susceptibility against cytoplasmic ribonuclease inhibitor, was highly catalytic and stable [34]. Currently, chemical modification and genetic engineering targeting Barnase to enhance its productivity and activity is a hot topic. Barnase has no disulfide bonds and thus it does not require any post-translational modifications, divalent cations or other non-peptide components. Hence due to these favorable features, Barnase is active in any cell type in which it is expressed [35]. Barnase efficiently cleaves target RNA, and this property has been exploited in a wide variety of biological applications since introduction of this enzyme into cells causes cell death. Specific ablation of particular cells is feasible by directing Barnase gene expression via the use of cell-specific promoters. Alternatively, proteins that target Barnase to specific cells endow specificity to Barnase action [36-38].
Major problems in human pancreatic-type RNases is that these are sensitive to inhibition by cytoplasmic ribonuclease inhibitor (RI) found in every mammalian cell studied so far [39]. While examining the susceptibility of Barnase to RI, it was found that Barnase is fortunately insensitive to inhibition by RI. Cancer has specific marker HER2 antigen overexpressed in human neoplasma, and HER2 expression causes the ovarian and breast cancinomas. In a previous study, it was reported that two Barnase molecules fused with a single-chain variable fragment (scFv) of the humanized antibody 4D5, recognizes the domain of HER2 and produced scFv 4D5-diBarnase [38]. An immunoRNase (IR) was created that included two ribonucleases had specific cytotoxicity limited by the cell surface density of the HER2 antigen. This configuration enabled the introduction of twice the ribonuclease activity into cells with just one HER2 receptor. The scFv 4D5-diBarnase was able to interact specifically with HER2- positive human ovarian cells, be internalized into the cells, and exerted cytotoxicity.
RNase Sa: Another promising microbial T1 family belonging to Streptomyces has representative members such as RNases Sa (strain BMK), Sa2 (strain R8/26) and Sa3 (Streptomyces aureofaciens, strain CCM 3239) which structurally resembled with each other and have identical amino acids in 48 out of 96 positions. Out of these three RNases, RNase Sa3 is potentially active and highly catalytic against cancer cells rather than RNase Sa2 and RNase Sa1 [40]. Although, RNase Sa3 tertiary structure is quite similar to RNase Sa2 but still RNase Sa3 is more cytotoxic towards malignant cells [41].
The cytotoxicity of RNase Sa3 depends on some structural elements and sequence motifs. The replacement of Asp and Glu residues with Lys on the surface of RNase Sa3 and variation in their charge form acidic to basic protein increases toxic effect against malignant cells. Due to reversing charges, it could generate sufficient cytotoxic effects in RNase Sa3 [40,42]. Cytotoxic activity of RNase Sa correlates with the change in its net charge from negative to positive. Site-directed mutation is one of approaches which allows the creation of charges on enzymes and there by produces toxic effect(s) with bare minimum side effects on the cells (30,43,44). At low concentration (IC50 μM), the RNase Sa3 has been shown to cause toxic effect towards human erythroleukemia K562 cells without being inhibited by the cytosolic RNase inhibitor. These malignant cell-selective characteristics revealed that RNase Sa3 could be used as a potential anticancer agent for treating leukemia [45].
α-Sarcin: The α-Sarcin is composed of 150-residues polypeptide toxin, while Mitogillin and Restrictocin are quite small (approximately 17 kDa) ribosome-inactivating proteins (RIPs) produced by the Aspergillus giganteus. α-Sarcin belongs to type 1 group of RIPs and are predominant members of the fungal Ribotoxins that display 3-D structure. His50, Glu96 and His137 residues are responsible for α-Sarcin catalytic activity towards malignant cells [46,47]. These three fungal ribotoxin act on specific RNA to accomplish the cleavage of phosphodiester bonds in the universally conserved α-Sarcin domain of 28S rRNA and thus inhibit the protein synthesis [19]. The α-Sarcin interacts with lipid bilayer of malignant cell membrane and fuses with the cell leading to inhibition of protein synthesis and induction of apoptosis [47,48]. The α-Sarcin has been previously reported as a potent cytotoxin that promotes apoptosis in human rhabdomyosarcoma cells. Researcher found that at molecular level ribotoxin belongs to super family of RNases.
Analysis of the Mitogillin gene and PCR-mediated site-specific mutagenesis suggested that positive domains in ribotoxin, which share homologies with motifs in ribosome-related proteins and liable to be targeting the ribotoxins to the ribosome. Due to this application, the ribotoxins are used as tools in research, therapeutics and as diagnostic agents [19,18,47]. The α-Sarcin is strongly cytotoxic against virus-infected mammalian cells or transformed cells without the use on any permeabilizing agent. The RNases differing from each other by peptides and proteins have also been isolated from several mushroom species. RNase form Pleurotus sajor-caju exerts anti-proliferative action on leukemia, hepatoma, and antimitogenic action on mouse spleen cells because of presence of ribosome-inactivating proteins. In past decade, H. marmoreus, Lyophyllum shimeiji, Calvatia caelata, F. velutipes and Pleurotus tuber-1 mushrooms have been used to extract ribosome-inactivating proteins [49,50].
Actibind and RNase T2: Actibind is a protein isolated from black mold Aspergillus niger which is a well-known microorganism used in bio- and food-industry. In previous finding, Actibind was found to control the cancer development by preventing the growth of malignant cells in mammalian cells [23,24]. RNase T2 was also found to bind actin in migrating cells in both animals and humans. RNase T2 enzyme prevents the blood supply to tumor cells while Actibind prevents the malignant cells to move through the blood stream to form new metastases [20,18]. The encouraging outcome was that the Actibind didn’t show any toxic effect on normal cells thus minimizing the possibility of side effects. In future, fungal Actibind and RNase T2 could be used as frontline therapies in the fight against cancers [19,18].
Ribonuclease P (RNase P): The RNase P is a type of endoribonuclease that targeting the t-RNA degradation at 5’end into smaller components and universally composed of both protein and RNA. RNase P enzyme occurs in all three domains of microbes i.e., bacteria, archaea and eukarya. Bacterial RNAse P differs from other two domains of microbes due to its protein composition and RNA structure. In vivo, bacterial RNase P has been associated with single protein [51]. RNAse P is a divalent cation-dependent endoribonuclease which acts as a riboenzyme and is encoded by protein subunit rnpA gene and RNA subunit rnpB [52].
RNase P functions to breakdown the precursor sequence of RNA on RNA molecules. The ongoing cancer therapy approaches have a major problem to differentiate between the cancer cells and the normal cells. This problem may be overcome by specific chimeric molecules, which are specific to the cancer cells, and thus efficiently act on the specific targets. RNase P has catalytic subunit M1 which catalyses the hydrolytic removal of 5´-leader sequence of t-RNA and also permits the direct gene targeting. M1 RNA subunit can be targeted to the mRNA by the addition of guide sequence at the 3´-terminal (M1-GS). M1 RNA becomes M1-GS which cleaves the mRNA and will thus halt formation of the fusion proteins, which are specific for the cancer cells [53,18].
Genetically engineered and chemical modified RNases: Microbial RNases have been conjugated or modified with a variety of molecules i.e. as ethylenediamine or polyethyleneimine to improve their cellular uptake and reduced affinity for RI. Microbial ribotoxin ‘Rrestrictocin’ bound to the antibodies resulted in an increase in the cytotoxicity of this construct for cancer cells while decreasing the systemic toxicity during coupling of RNase. Various conjugates of RNases and cell targeting moieties such as transferrin or antibodies against the transferrin receptor or CD22 have been constructed which in part dramatically increased the cytotoxicity of the RNases [54]. These modifications in RNase cause cellular uptake into cancer cells with Columbic interaction [55-57]. Although, scientists observed that the catalytic activity was decreased by cationization of the carboxyl groups modification which endowed the proteins with incredible cytotoxicity. Poly[N-(2- hydroxypropyl)methacrylamide] molecules conjugate with BS-RNase or RNase A produced active variants for targeting malignant cells in a mouse model [58]. According to these observations, microbial RNase may yield more active variants against malignant cells without interacting with RI molecule in the cells [59].
Another finding observed by scientists was about the RI binding and the change of cytotoxic nature by producing mutants or mutation in base pair residues or change in structure of enzymes [40,60]. In some cases, the combination of catalytic active region and RI evasion, the active segment combines to produce new active molecule with more advantages forms of RNases known as chimers [59]. RNase1 and Onconase were used to produces chimers resulting in good catalyitic activity and RI evasion properties. These techniques we can produce active microbial RNases with effective and efficient anticancer outcomes [61].
Cytotoxicity Mechanisms of Microbial RNases towards Cancer Cells
How and why do microbial RNases and other RNases preferentially kill malignant cells? Malignant cells have been endorsed with numerous characteristics associated with cytotoxic pathway like selective sensitive RNA hydrolysis, intracellular routing and selective membrane recognition by receptor or ligand. These few postulates help us to understand the RNases internalized pathways into malignant cells. Microbial RNases may selectively recognize and hence get internalized by malignant cells due to the molar ratio of ethanolamine phospholipids to choline phospholipids in the plasma membrane which appeared to increase after neoplastic transformation. This outcome results in an increase in the anionic content of the membrane that thus facilitates the adsorption of RNases. Another alternative explanation for the preferential selection of cancer cells could rely on a different intracellular route for internalization, only present in malignant cells but not in normal cells. Trans-Golgi network present only in malignant cells may leads to a precise organelle allowing the protein to translocate to the cytosol in case of BS-RNase [62].
According to this observation, the malignant cells might be more sensitive towards RNase internalization pathways than normal cells which eventually laeds to the toxic effects of RNA hydrolysis (Figure 2). It has been shown that the cells with inactivated tumour suppressor genes may activate proapoptotic pathways that cannot be accessed in normal cells [11]. A scheme of potential interactions of cytotoxic RNase with host cellular components. RNase binds on to the cell membrane by lipid rafts or receptor (specific or nonspecific interactions), enters into the cytosol by endocytosis and targets the cellular RNA causing degradation of RNA into nucleotides, which leads to alteration or blocking of the protein synthesis. Blockage of protein synthesis helps to activate the Caspases-driven cell death (apoptosis or necrosis).
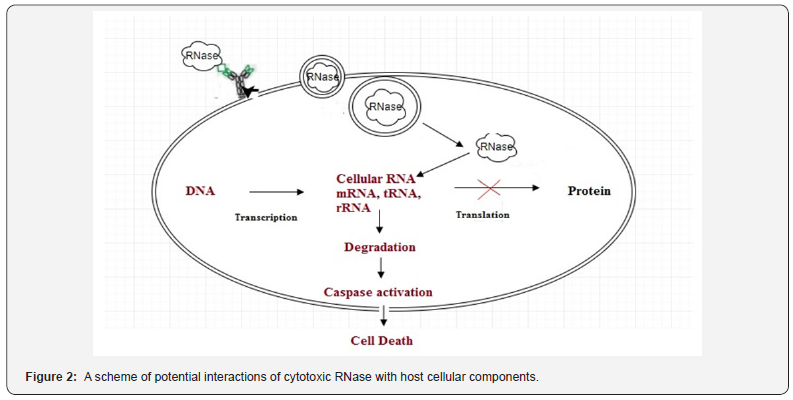
RNase-induced cell death is a complex, tightly controlled and multistep process for which several pathways and possible mechanisms of different RNases have been suggested such as;
A. The cytotoxic effects of microbial RNase are associated with catalytic cleavage of cellular RNA including t-RNA, rRNA, mRNA and the non-coding RNA (microRNAs) to inhibit gene expression [15].
B. Non-catalytic electrostatic interaction of exogenous enzyme with cell components.
C. Modulation of the membrane calcium-dependent potassium channels and ras-oncogene functions.
D. Neutralizing action of the cytosolic RNase inhibitor and the regulation of complicated pathways.
Before the internalization, extracellular RNase interacts with the surface of target cell by membrane lipids, ion channels, receptors and by non-specific electrostatic binding on the cell surface. Membrane proteins as well as lipids both are involved in interaction with RNase specifically. Cytotoxicity of microbial RNases has been observed due to their cationic nature. Mechanism of Binase internalization into transformed cells was based on the positive charge which allows it to bind anionic groups on the surface of tumor cells [40]. Malignant cells express more acid phospholipids on the outer surface of their membrane than their non-tumor surfaces, and therefore are more negatively charged [63]. Based on this argument, scientists demonstrated earlier that the cytotoxicity of RNase from Streptomyces aureofaciens (RNase Sa) is enhanced to a great extent by replacement of negative charges on the surface of the molecule by positively charged residues [44,64]. The cytotoxicity of RNase Sa was affected by charge reversal mutants that correlates with their positive charge, although the activity and stability were the same. Besides, RNase Sa2 and RNase Sa3 are isoenzymes of S. aureofaciens which acquired higher positive charge than that of Sa (pI 5.3 and 7.2, correspondingly) and both RNases toxicity correlated with their charges [64].
Some scientists demonstrated that chemical modification in charge molecules leads to increase in the positive charge on RNase and also cell-binding ability appeared to be enhanced by chemical modification. RNase A and RNase 1 have been observed to be more cytotoxic and cell-binding towards malignant cells because of increase in net positive charges [9]. In previous studies there has been an evidence for existence of non-protein receptor-like sites on target cell surface. The RNase enters into cells by adsorption- mediated endocytosis by sulfhydryldisulfide of Globo H or lipid molecule interaction [65]. According to some authors, RNase A variant and HP-RNases are internalized in A4310r K562 cells by endocytosis fluid-phase uptake process [66,67].
A Look at The Microbial Ribonuclease-Receptor Interaction Mechanisms
RNases are known as RNA-hydrolyzing enzymes that exert numerous biological effects exogenously for degradation of cellular RNA apart from performing their main functions in the cell. Other RNase like as RNase A and RNase 1 bind with the surface of cancer cells due to acidic lipid raft, heparin sulphate and other glycans exposed on the cell surface(s) and non-malignant cells [68]. Because of unusual pattern of tumorassociated carbohydrate antigen (TACAs) present uniquely in cancer cell surface may act as potential acceptors for exogenous RNases. These molecules create more anionic surfaces on malignant cells than a normal cell [68-70]
Secondly, Globo H is a neutral hexasaccharide glycosphingolipid located endogenously on the outer membrane of epithelial cells of mammary, uterine, gut, kidney and pancreas which acts as receptor/ ligand for RNase1 [68,71]. The Globo H, sialic acid and heparin sulphate play vital roles in the cellular uptake of RNase due to increasing anionic nature of cancerous cell surface. RNases cationic side-chain interacts with glycan or sialic acid or lipids of cancerous cell-surface with non-specific Columbic interactions. Globo H has been reported as a more specific ligand for RNAse 1 than RNase A. Hyaluronic acid (HA)- RNase A is a nano-complex with cationic lipid molecules which inhibits the cancer proliferation. RNase A binds with HA due to supramolecular interaction with carrier lipidoids facilitates cancer cell targeting via interaction with over expressed CD44. HA-RNase A can specifically bind to the CD44 receptor through Columbic interactions leading to initiation of endocytosis pathways for cellular uptake of RNase [72]. These nanocomplexes are also used for drug delivery to the cancer cells. In previous studies, HA-RNase A was efficiently delivered to the CD44-overexpressing A549 cells which extensively inhibited cancer cells proliferation. This provided an efficient method of targeted cancer cell therapy [73].
Virus T antigen (TAg) also known as an oncoprotein is exposed on the surface of MLE12 pneumocytes. TAg is covalently bound with RNA via a phosphodiester bond between 5’-phosphate and the β-hydroxyl group of a serine residue, and this complex acts as a potent receptor for RNase [74]. The RNase degraded the RNA which was associated with TAg oncoprotein of transformed mouse cells into ribonucleoside phosphates. Binase binds with TAg oncoprotein, which is present in MLE12 pneumocytes membrane as receptor and such binding causes hydrolyzes of TAg-associated RNA. In MLE12 pneumocytes, the Binase exerts a cytotoxic effect even without its internalization while Binase remained insensitive to non-transformed type II pneumocytes cell lines [72].
Internalization of RNase from Bilayer Membrane and Cytosol
After binding to target cell surface, the cytotoxic RNase is internalized by endocytosis. Cytotoxic RNases have been localized in endosome but in some RNases, the cytotoxicity is blocked by some energy-dependent process. Endocytic mechanism is still scarcely understood but other RNases like Onconase and RNase A follow Clathrin and Dynamin-independent pathways. RNases enzymes transferred faster and were more toxic to malignant and non-malignant cells by endocytosis process [75]. RNase transportation from extracellular surface of malignant cells to cytosol follows two different approaches for RNase-associated toxicity:
A. Drugs disrupt the intercellular transport of cytotoxicity of RNase and leads to intercellular trafficking may account for indirect consequences on cellular pathway in cancer cell lines [42,75].
B. Fluorescence label with specific marker for organelles to trace the intercellular route and approach implemented on Onconase and BS-RNase for tracking intercellular route in the target cells was reported. RNases evade the cell membrane via charge based molecule, non-protein receptor-dependent mechanisms or endocytosis or direct translocation. β-Amyloid proteins, Prions, Calcitonin and unchaperoned positively charged molecules are able to directly translocate the RNase to the cytoplasm or induce membrane damage and cell malfunction mediated by ion channel formation [40,76]. The α-Sarcin molecules translocate into the cytosol by artificial lipid vessels that involved internalization through acidic endosomes in in vivo experiment. Endocytosis-mediated acidic vessel is responsible for internalization of RNases form extracellular matrix to the cytoplasm. For α-Sarcin, the internalization mechanism appeared to be Clathrin-independent. While the cytotoxicity caused by Onconase and G88R RNase A is Dynamin-independent [47,75,76].
C. Cellular RNA Degradation By Microbial RNase In Cytosol: Microbial RNase are involved the degradation of the cellular RNAs, their transport from endosomes to the Golginetwork and the endoplasmic reticulum [62]. RNases have been shown to harbour ribonuclease activity in cytosol by their chemical modification or addition of residues through site-directed mutation(s) for greater stability of enzymes in cytosol environment. RI protein acts as a safeguard against exogenous RNase and is found in cytosol [75]. Microbial RNases have some residues which are sensitive to RI binding that prompts the blocking or inhibition of the ribonucleolytic activity of the RNase in the cytosol. The human cells RI protein is however, not sensitive to bacterial RNases. Onconase evades RI action in vivo causing intense cytotoxicity but RI molecules being partially silenced by BSRNase has been reported because their dimeric forms are maintained by non-covalent interactions in the reducing environment of cytosol which are probably not inhibited by RI [77].
RNase progression from the endosome compartment to the Golgi complex was found in tumour cells but not in the normal cells [75,78]. The cytotoxic activities of RNase 1 and α-Sacrin severely disrupted the retrograde transport from the Golgi complex to ER, Angiogenin and RNase 1 [75]. RNases once translocated to the cytosol from the pre-ER compartment degrade cellular RNA and nuclear RNA. The signal recognition particles, which target proteins to ER, contain RNA as a component of highly conserved RNA-protein core [40]. Hydrolysis of small non-RNA into micro- RNA causes the alteration of gene expression and thus causes the killing of malignant cells. Binase enzyme interaction with ionic pathways might be involved in the cell proliferation control and appearance of cell phenotypes. These interaction of Binase with the ionic pathways may block Ca2+-activated K+ channels and thus inhibits the proliferation of ras-transformed fibroblasts without any effect on the normal cells, and in cells-transformed by src or fms oncogenes [ 40].
Thus, ras oncogene of expressing cells was more sensitive to Binase than the non-expressing cells. Likewise, Onconase exhibited cytotoxic activity towards ras-transformed mouse fibroblasts. These data suggested that ras-targeting RNases and ras proteins provide therapeutic possibilities for cancer therapy [79]. Degradation of cellular RNA by RNases arrests protein synthesis as well as induces apoptosis in the affected cells. In case of Onconase, there are two lines of evidences which triggered apoptosis i.e., cytotoxic and cytostatic effect [75]. Both effects of Onconase were observed apparently after 24-48 h of drug administration in the treated cells. Cylcoheximide or emetine was used for rapid inhibition of protein synthesis within 2-4 h of incubation period. The growth of human histocytic lymphoma U93784 and leukaemia HL6019 cell lines was arrested by Onconase at G1/S checkpoint of the cellular cycle [80]. The degradation of RNA can trigger apoptotic pathways in malignant cells. They also observed that besides degrading t-RNA, micro-RNA or small interfering RNA produced from degraded RNA can play roles in specific cell regulation [15]. Above studies clearly illustrated that the enzyme-RNA interaction as well as other direct and mediated effects of cytotoxic RNases are important for understanding the mechanisms of RNase cytotoxicity towards malignant cells [19,76]. These information help the scientists to identify cellular targets of cytotoxic RNases as well as distinguishes between their direct and indirect effects of ribonucleolytic action. Caspase-dependent process, low molecular weight compounds, alteration in protein and NF-κB signal pathway are cell death mechanisms which involve in response to cytotoxic RNases. In a previously study, Onconase decreased the NF-κB1 transcription factor in pleural mesothelioma cells and restrained the canonical NF-κB dependent pathways. Binase-induced cell death reduced the mitochondrial potential and ligand-dependent apoptosis in Kasumi-1 and B-16 cells [32,80]. Mitochondrial was potential reduced by Binase via formation of mitochondrial pores which activated the caspase 8 for increased Ca2+ level and a decline in reactive oxygen species was observed. The tumor necrosis factor (TNF) present on B-16 cells surface increased the response to Binase but TNF increased such a response by a factor of 16 in Kasumi-1 cells [32].
In another study, the genes of canonical NF-κB dependent signal pathway and pro-inflammatory caspase 1 and 4 gene were activated by Binase. Binase showed the cytotoxic effect against cancers cell line by activating the TNF through caspase 8 and NF-κB which further activated caspase 3, 4 and 7 leading to cell death [32,80]. Currently, we are still unclear which of these targets is more effective in triggering apoptotic process and thus more promising for potent cancer therapy. The unraveling of the underlying role of cytotoxic RNases to target the cellular RNA in cancer/ tumour cells by untraceable routes and pathways leading to cell death is still incomplete. In future, the RNases may prove to be promising alternatives to develop antitumour drugs with extended therapeutic applications [80-88].
Conclusion
The killer strategy of microbial RNases has highlighted their extended use as antitumour drugs or therapeutic agents to eliminate the tumour cells. Microbial RNases follow binding of a main target, cellular RNA, selective intracellular routing and membrane specific recognition processes. The cytotoxic pathways of RNases serve as working platforms for the creation of new anticancer drugs in future. Now, when major biological activities of microbial RNases have been exposed, time has come to translate this knowledge into new therapeutic applications. Little technique, chemical modification in ligand/ receptor or mutation may lead to an increase in the cytotoxic properties of microbial RNase molecules. Such efficient RNases may selectively allow the cytotoxic pathway to work in the malignant cells. The chemical modification(s) of known RNases to enhance their resistance to intracellular inhibitors may yield more potent long-lasting RNases. The creation of new ribozyme-type highly specific RNases aimed to precise destruction of the products of oncogenes is another approach for anticancer therapeutics. Further development in RNase studies will be prerequisite for development of effective drug against cancer cells and anticancer therapeutics.
Acknowledgment
The authors are thankful to University Grants Commission, Government of India, New Delhi for the financial support to one of the authors Mr. Rakesh Kumar in the form of a RGNF-SRF sanctioned vide a Letter No. F1-17.1/2013-14/RGNF-2013-14- SC-HIM-51265, as well as the Department of Biotechnology, Ministry of Science & technology, Government of India for providing financial support to the parent department. Further, the authors declare that no conflict of interest exists among themselves or with their parent institution.
References
- Jones W (1920) The action of boiled pancreas extract on yeast nucleic acid. Am J Physiol 52: 203-2017.
- Rajashree A, Deshpande, Shankar V (2002) Ribonucleases from T2 Family Critical Reviews in Microbiology 28(2): 79-122.
- Noguchi J (1924) On the decomposition of nucleic acids through takadiastase. Biochem Z 147-255.
- Sato K, Egami F (1957) Studies on ribonucleases in takadiastase. J Biochem 44(11): 753.
- Nicholson AW (1999) Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol Rev 23(3): 371-390.
- Conrad C, Rauhut R (2002) Ribonuclease III: new sense from nuisance. Int J Biochem Cell Biol 34(2): 116-129.
- Condon C, Putzer H (2002) The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res 30(24): 5339-5346.
- Aravind L, Koonin EV (2001) A natural classification of ribonucleases. Methods Enzymol 341: 3-28.
- Futami J, Maeda T, Kitazoe M, Nukui E, Tada H, et al. (2001) Preparation of potent cytotoxic ribonucleases by cationization: enhanced cellular uptake and decreased interaction with ribonuclease inhibitor by chemical modification of carboxyl groups. Biochemistry 40(25): 7518- 7524.
- Smith MR, Newton DL, Mikulski SM, Rybak SM (1999) Cell cyclerelated differences in susceptibility of NIH/3T3 cells to ribonucleases. Exp Cell Res 247(1): 220- 232.
- Iordanov MS, Ryabinina OP, Wong J , Newton DL , Rybak SM, et al. (2000) Molecular determinants of apoptosis induced by the cytotoxic ribonuclease onconase: evidence for cytotoxic mechanisms different from inhibition of protein synthesis. Cancer Res 60(7): 1983-1994.
- Prior TI, Kunwar S, Pastan I (1996) Studies on the activity of barnase toxins in vitro and in vivo. Bioconjug Chem 7(1): 23- 29.
- Matousek J (2001) Comp Biochem Physiol C Toxicol Pharmacol 129(3): 175-191.
- Vescia S, Tramontano D, Augusti-Tocco G, D’Alessio G (1980) In vitro studies on selective inhibition of tumor cell growth by seminal ribonuclease. Cancer Res 40(10): 3740-3744.
- Ardelt W, Ardelt B, Darzynkiewicz Z (2009) Ribonucleases as potential modalities in anticancer therapy. Eur J Pharmacol 625(1-3): 181-189.
- Belyaeva MI, Kyune MF, Nuzhina AM (1964) Effect of bacterial deoxyribonuclease on Ehrlich’s ascites carcinoma cells in vitro. Fed Proc Transl Suppl 23: 345-348.
- Makarov AA, Kolchinsky A, Ilinskaya ON (2008) Binase and other microbial RNases as potential anticancer agents. Bioessays 30(8): 781-790.
- Shlyakhovenko VO (2016) Ribonucleases: Possible new approach in cancer therapy. Exp Oncol 38(1): 2-8.
- Suri S, Panda B, Javed S, Mohd A (2007) RNase: a novel enzyme for treatment of cancers. Internet J Oncol. 5(1): 1-5.
- MacIntosh GC (2011) RNase T2 Family: Enzymatic Properties, Functional Diversity, and Evolution of Ancient Ribonucleases. In: Nicholson AW (ed.) Ribonucleases 26: 89-114.
- Maquat LE, Arraiano C (2008) RNA turnover in bacteria, archaea and organelles. Acad Press 447: 620.
- Yuki S, Kondo Y, Kato F, Kato M, Matsuo V (2004) Noncytotoxic ribonucle¬ase, RNase T1, induces tumor cell death via hemagglutinating vi¬rus of Japan envelope vector. Eur J Biochem 271(17): 3567-3572.
- Yoshida H (2001) The ribonuclease T1 family. Methods Enzymol 341: 28-41.
- Carreras SN, Alvarez GE, Herrero GE, Tome J, Lacadena J, et al. (2008) The therapeutic potential of fungal ribotoxins. Curr Pharm Biotechnol 9(3):153-60.
- Aphanasenko GA, Dudkin SM, Kaminir LB, Leshchinskaya IB, Severin ES (1979) Primary structure of ribonuclease from Bacillus intermedius 7P. FEBS Lett 97(1): 77-80.
- Polyakov KM, Lebedev AA, Okorokov AL, Panov KI, Schulga AA. et al. (2002) The structure of substrate-free microbial ribonuclease Binase and of its complexes with 3′GMP and sulfate ions. Acta Crystallogr D Biol Crystallogr 58: 744-750
- Ilinskaya ON, Zelenikhin PV, Petrushanko IY, Mitkevich VA, et al. (2007) Binase induces apoptosis of transformed myeloid cells and does not induce T-cell immune response Biochem. Biophys Res Commun 361(4): 1000-1005.
- Cabrera-Fuentes HA, Kalacheva NV, Mukhametshina RT, Zelenichin PV, Kolpakov AI, et al. (2012) Binase penetration into alveolar epithelial cells does not induce cell death. Biochemistry (Moscow) Supplement Series B: Biomed Chemistry 6(4): 317-321
- Mitkevich VA, Makarov AA, Ilinskaya ON (2014) Cell targets of antitumor ribonucleases. Mol Bio 48(2): 181-188.
- Mitkevich VA, Petrushanko IY, Spirin PV, Fedorova TV, Kretova OV, et al. (2011) Sensitivity of acute myeloid leukemia Kasumi-1 cells to binase toxic action depends on the expression of KIT and AML1-ETO oncogenes. Cell Cyclem 10(23): 4090-4097.
- Mitkevich VA, Kretova OV, Petrushanko IY Burnysheva KM, Sosin DV, et al. (2013) Ribonuclease binase apoptotic signature in leukemic Kasumi-1 cells. Biochimie 95(6): 1344-1349.
- Cabrera-Fuentes HA, Aslam M, Saffarzadeh M, Kolpakov A, Zelenikihin P, et al (2013) Internalization of Bacillus intermedius ribonuclease (BINASE) induces human alveolar adenocarcinoma cell death . Toxicon 69: 219-226.
- Balandin TC, Edelweiss E, Andronova NV, Treshalina EM, Sapozhnikov AM Deyev, (2009) Antitumor activity and toxicity of anti-HER2 immunoRNase scFv 4D5-dibarnase in mice bearing human breast cancer xenografts. Invest New Drugs 29(1): 22-32.
- Hartley RW (1989) Barnase and barstar: two small proteins to fold and fit together. Trends Biochem Sci 14(11): 450-454.
- Prior TI, FitzGerald DJ, Pastan I (1992) Translocation mediated by domain II of Pseudomonas exotoxin A: transport of barnase into the cytosol. Biochemistry 31(14): 3555-3559.
- Prior TI, Kunwar S, Pastan I (1996) Studies on the activity of barnase toxins in vitro and in vivo. Bioconjug Chem. 7(1): 23-29.
- Deyev SM, Waibel R, Lebedenko EN, Schubiger A, Pluckthun A (2003) Design of multivalent complexes using the barnase*barstar module. Nat Biotechnol 21(12): 1486-1492.
- Haigis MC, Kurten EL, Raines RT (2003) Ribonuclease inhibitor as an intracellular sentry. Nucleic Acids Res 31: 1024-1032.
- Makarov AA, Ilinskaya ON (2003) Cytotoxic ribonucleases: molecular weapons and their targets. FEBS Lett 540(1-3): 15-20.
- Pace CN, Hebert EJ, Shaw KL Schell D, Both V, Krajcikova D, et al. (1998) Conformational stability and thermodynamics of folding of ribonucleases Sa, Sa2 and Sa3. J Mol Biol 279(1): 271-286.
- Trevino SR, Scholtz M, Pace CN (2007) Amino acid contribution to protein solubility: Asp, Glu and Ser contribute more favourably than the other hydrophilic amino acids in RNase Sa. J Mol Biol 366(2): 449- 460.
- Hlinkova V, Urbanikova L, Krajcikova D, Sevcik J (2001) Purification, crystallization and preliminary X-ray analysis of two crystal forms of ribonuclease Sa3. Acta Crystallogr D Biol Crystallogr 57(5): 737-739.
- Ilinskaya ON, Dreyer F, Mitkevich VA, Shaw KL, Pace CN, Marakov AA (2002) Changing the net charge from negative to positive makes ribonuclease Sa cytotoxic. Protein Sci 11(10): 2522-2525.
- Sevcik J, Urbanikova L, Leland PA, Raines RT (2002) X-ray structure of two crystalline forms of a streptomycete ribonuclease with cytotoxic activity, J Biol Chem 277(49): 47325-47330.
- Kao R, Davies J (1995) Fungal ribotoxins: a family of natu¬rally engineered targeted toxins? Biochem Cell Biol. 73(12): 1151-9.
- Olmo N, Turnay J, Buitrago GG, et al. (2001) Cytotoxic mechanism of the ribotoxin -sarcin induction of cell death via apoptosis. Eur J Biochem 268: 2113-2123.
- Carreras SN, Tome AJ, Garcia OL, Batt CA, Onaderra M (2012) Production and characterization of a colon cancer-spe¬cific immunotoxin based on the fungal ribotoxin α-sarcin. Protein Eng Des Sel 25(8): 425-435.
- Ng TB (2004)Peptides and proteins from fungi. Peptides 25(6):1055- 1073.
- Xu X, Yan H, Chen J, Zhang X (2011) Bioactive proteins from mushrooms. Biotech Adv. 29(6): 667-674.
- Kouzuma Y, Mizoguchi M, Takagi H, Fukuhara H, Tsukamoto M (2003) Reconstitution of archaeal ribonuclease P from RNA and four protein components. Biochem Biophys Res Commun 306(3): 666-673.
- Marquez SM, Chen JL, Evans D, Pace NR (2006) Structure and Function of Eukaryotic Ribonuclease P RNA. Mol Cell 24(3): 445-456.
- Cobaleda IC, Garcia IS (2009) In vivo inhibition by a site-specific catalytic RNA subunit of RNase P designed against the BCR-ABL oncogenic products: a novel approach for cancer treatment. Blood 95(3): 731-737.
- Rybak SM, Newton DL (1999) Natural and engineered cytotoxic ribonucleases: therapeutic potential. Exp Cell Res 253(2): 325-335.
- Mitchell DJ, Kim DT, Steinman L, Fathman CG, Rothbard JB (2000) Polyarginine enters cells more efficiently than other polycationic homopolymers. J Pept Res 56(5): 318-325.
- Futami J, Maeda T, Kitazoe M, Nukui E, Tada H et al. (2001) Preparation of potent cytotoxic ribonucleases by cationization: enhanced cellular uptake and decreased interaction with ribonuclease inhibitor by chemical modification of carboxyl groups. Biochemistry 40(25): 7518- 7524.
- Futami J, Kitazoe M, Maeda T, Nukui E, Sakaguchi M (2005) Intracellular delivery of proteins into mammalian living cells by polyethyleniminecationization. J Biosci Bioeng 99(2): 95-103.
- Pouckova P, Skvor J, Gotte G, Vottariello F, Slavik JT et al. (2006) Some biological actions of PEG-conjugated RNase A oligomers. Neoplasma 53(1): 79-85.
- Arnold U Holfman UR (2006) Natural and engineered ribonucleases as potential cancer therapeutics. Biotechnol Lett. 28(20): 1615-1622.
- Leland PA, Raines RT (2001) Cancer chemotherapy-ribonucleases to the rescue. Chem Biol 8(5): 405-413.
- Boix E, Wu Y, Vasandani VM, Saxena SK, Ardelt W, et al. (1996) Role of the N-terminus in RNase A homologues: differences in catalytic activity, ribonuclease inhibitor interaction and cytotoxicity. J Mol Biol 257(5): 992-1007.
- Bracale A, Spalletti CD, Mastronicola M, Castaldi F, Mannucii L, et al. (2002) Essential stations in the intracellular pathways of cytotoxic bovine seminal ribonuclease. Biochem J 362(3): 553-560.
- Ran S, Downes A, Thorpe PE (2002) Increased exposure of anionic phospholipids on the surface of tumor blood vessels. Cancer Res 62(21): 6132-6140.
- Ilinskaya ON, Koschinski A, Mitkevich VA, Repp H, Dreyer F, et al. (2004) Cytotoxicity of RNases is increased by cationization and counteracted by KCa channels. Biochem Biophys Res Comm 314(2): 550-554.
- Bracale A, Castaldi F, Nitsch L, D’Alessio G (2003) A role for the intersubunit disulfides of seminal RNase in the mechanism of its antitumor action. Eur J Biochm 270(9): 1980-1987.
- Haigis MC, Kurten EL, Abel RL, Raines RT (2002) KFERQ sequence in ribonuclease A-mediated cytotoxicity. J Biol Chem 277: 11576 -11581.
- Bosch M, Benito A, Ribo M, Puig T, Beaumelle B, et al. (2004) A nuclear localization sequence endows human pancreatic ribonuclease with cytotoxic activity. Biochem 43(8): 2167-2177.
- Eller CH, Chao TY, Singarapu KK, Ouerfelli O, et al. (2015) Human cancer antigen Globo H is a cell-surface ligand for human Ribonuclease 1. ACS Cent Sci 1(4): 181-190.
- Nakano M, Kakehi K, Tsai MH, Lee YC (2004). Detailed structural features of glycan chains derived from α1-acid glycoproteins of several different animals: The presence of hypersialylated, Oacetylated sialic acids but not disialyl residues. Glycobiology 14(5): 431-441.
- Danishefsky SJ, Shue YK, Chang MN, Wong Ch (2014) Development of Globo-H Cancer Vaccine. ACC Chem Res 48(3): 643-652.
- Camejo EH, Rosengren B, Sartipy P, Elfsberg K, Camejo G, et al. (1999) CD44, a cell surface chondroitin sulfate proteoglycan, mediates binding of interferon-g and some of its biological effects on human vascular smooth muscle cells. J Biol Chem 274(27): 18957-18964
- Wang X, Li Y, Li Q, Neufeld CI, Pouli D, et al. (2017) Hyaluronic acid modification of rnase a and its intracellular delivery using lipid-like nanoparticles. J Control Release 263: 39-45.
- Mitkevich VA, Burnysheva KM, Ilinskaya ON, Pace CN, Marakov AA (2014) Cytotoxicity of RNase Sa to the acute myeloid leukemia Kasumi-1 cells depends on the net charge. Oncoscience 1(11): 738- 744.
- Carroll RB, Samad A, Mann A, Harper J, Anderson CE (1988) RNA is covalently linked to SV40 large T antigen. Oncogene 2(5): 437-444.
- Benito A, Ribo M, Vilanova M (2005) On the track of antitumour ribonucleases. Mol BioSyst 1: 294-302.
- Kourie JI, Henry CL (2002) Ion channel formation and membranelinked pathologies of misfolded hydrophobic proteins: the role of dangerous unchaperoned molecules. Clin Exp Pharmacol Physiol 29(9): 741-753.
- Kobe B, Deisenhofer J (1996) Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease. A J Mol Biol 264(5’): 1028-1043.
- Futami J, Nakui E, Maede T, et al. (2002) Optimum modification for the highest cytotoxicity of cationized. 132(2): 223-228.
- Gazdar AF, Minna JD (2001). Targeted therapies for killing tumor cells. Proc Natl Acad Sci USA 98(18): 10028-10030.
- Ardelt B, Juan G, Burfeind P, Salomon T, Wu JM, et al. (2007) Onconase, an anti-tumor ribonuclease suppresses intracellular oxidative stress. Int J Oncol 31(3): 663-669.
- Darzynkiewicz Z, Carter SP, Mikulski SM, Ardelt WJ, Shogen K (1988) Cytostatic and cytotoxic effects of Pannon (P-30 Protein) a novel anticancer agent. Cell Tissue Kinet 21(3): 169-182.
- Garipov AR, Nesmelov AA, Cabrera-Fuentes HA, Ilinskaya ON (2014) Bacillus intermedius ribonuclease (BINASE) induces apoptosis in human ovarian cancer cells Toxicon 92: 54-59.
- Heisterkamp N, Jenster G, Hoeve JT, Zovich D, Pattengale PK, et al. (1990) Acute leukemia in BCR/ABL transgenic mice. Nature. 344:251- 3.
- Madan T, Arora N, Sarma PU (1997) Ribonuclease activity dependent cytotoxicity of Asp fl, a major allergen of A. fumigatus. Mol Cell Biochem 175(1-2): 21-27.
- Olson BH, Jennings JC, Roga V, Junek AJ, Schuurmans DM (1965) Alpha sarcin, a new antitumor agent. Appl Microbiol. 13:322-326
- Otani H (1935). The mold enzymes splitting nucleic acid, Acta Schol Med Univ Imp Kioto 17:323.
- Sinatra F, Callari D, Viola M, Longombardo MT, Patania M, et al. (2000) Bovine seminal RNase induces apoptosis in normal proliferating lymphocytes. Int J Clin Lab Res 30(4): 191-196.
- Vescia S, Tramontano D, Augusti-Tocco G, D’Alessio G (1980) In vitro studies on selective inhibition of tumor cell growth by seminal ribonuclease. Cancer Res 40(10): 3740-3744.
- ZelenikhinPA, Kolpakov AI, Cherepnev GV, Ilinskaia ON (2005) Induction of apoptosis of tumor cells by Binase. Mol Biol (Mosk) 39(3): 457-463.






























