In Silico Characterization of Various Steps of Validamycin Pathway Using Gene Annotation and Protein Fold Based Selection Approach
Ruchika Bhat1,2,3, Rukmankesh Mehra1,4 and Amit Nargotra1,5*
1Discovery Informatics Division, CSIR-Indian Institute of Integrative Medicine, India
2Department of Chemistry, Indian Institute of Technology, India
3Supercomputing Facility for Bioinformatics and Computational Biology, Indian Institute of Technology, India
4School of Biotechnology, University of Jammu, India
5Academy of Scientific and Innovative Research, CSIR-Indian Institute of Integrative Medicine, India
Submission: October 06, 2017; Published: November 27, 2017
*Corresponding author: Amit Nargotra, Discovery Informatics, CSIR-Indian Institute of Integrative Medicine, Canal Road, Jammu 180001, India, Tel: 0191-2569021; Email: ruchikabhat31@gmail.com
How to cite this article: Ruchika B, Rukmankesh M, Amit N. In Silico Characterization of Various Steps of Validamycin Pathway Using Gene Annotation and Protein Fold Based Selection Approach. Adv Biotech & Micro. 2017; 7(3): 555715. DOI : 10.19080/AIBM.2017.07.555715
Abstract
Streptomyces hygroscopicus subsp. jinggangensis 5008, the major producer of anti-fungal aminoglycoside antibiotic jinggangmycin/ validamycin (VAL-A), is used to control sheath blight disease of rice plant for more than 40 years. Because of its high level of effectiveness as a fungicide as well as its lack of threat to human and animal health, validamycin has been one of the most important and widely used plant protection reagents against fungus Pellicularia sasakii (Rhizoctonia solani) in East Asia. In this work, S. hygroscopicus genome has been analyzed particularly with respect to the gene cluster responsible for the production of validamycin to enhance the information about its biochemical pathway, using in silico approach. Gene prediction was carried out for the 45kb long genome part reported to be involved in biosynthesis using GeneMark S tool. The research work was aimed at associating the genes/enzymes involved at each step of validamycin synthesis pathway, by the application of protein fold based selection of substrates approach. All the nine reaction steps involved in the biochemical pathway were annotated and the results were confirmed using reported annotated four steps. This work highlights the significance of an in silico prediction approach towards annotating novel biochemical pathways
Keywords: Validamycin; S. hygroscopicus; Gene prediction; Protein fold based selection; Biochemical pathway and protein folds
Highlights
A. Homology based annotation of genes for characterization of validamycin pathway steps.
B. Identification of probable enzymes working at each step by classifying genes according to known enzyme classes.
C. 3D structure prediction of probable enzymes of the pathway, their validation and the identification of protein folds.
D. Docking studies for the generation and selection of enzyme-substrate complexes.
E. Stability analysis of the proposed substrates-enzyme complexes using MD simulation.
Introduction
Validamycin A (synonyms validamycin, jinggangmycin, VAL-A), which is an antifungal agent, is biosynthesized by the species Streptomyces hygroscopicus var. jinggangensis 5008, S. hygroscopicus var. limoneus and S. hygroscopicus 10-22 [1-4]. It is used as crop protectant against fungal action of Rhizoctonia solani [2,5]. The mechanism of action of this antifungal drug lies in the inhibition of trehalase enzyme. In fungi, trehalose is a common storage carbohydrate and the enzyme trehalase hydrolyzes it to produce glucose, thus providing energy for the necessary cellular mechanism of fungus. Due to the inhibition of trehalase enzyme, fungus cells are deprived of glucose and eventually die off starving [6-8].
Validamycin belongs to the large family of sugar-derived microbial secondary products. There are various other C7N aminocyclitols natural products secondary metabolites like pyralomicin (used as antibiotics), cetoniacytone (an antitumor agent) and validamycin [9]. Based on the fact that the initial step of cyclization of sedoheptulose-7-phosphate to give 2-epi-5-epi- valiolone remains common, with further incorporation of 2-epi- 5-epi-valiolone into various secondary metabolites like acarbose (which is used as an anti-diabetic drug) and validamycin (an antifungal drug), some similarity in their biochemical pathways have been suggested [9-11]. Compounds like 2-epi- 5-epi-valiolone, 5-epi-valiolone, valienone, and validone on the basis of experimentation carried out using isotopically labeled precursors, were reported by Floss and co-workers as the intermediates of biosynthetic machinery of validamycin [4,10].
Bioinformatics through its tools and data sets has helped the researchers in various ways to reveal biological mysteries at the gene and protein level. Genome annotation, structural organization, functional proteomics etc. are now a part of exploratory science and are used to answer major biological problems. The data provided by various bioinformatic databases regarding protein folds and domains help in efficiently understanding the biological role of an enzyme, based on which we can predict their mode as well as site of action. Although this won't be wrong to say that bioinformatics just gives a probabilistic solution and in vivo experimentations prove it, but the time saved to screen out the neighborhood path for the solution makes the in silico approach appropriate for providing a rational approach to biological research [12-14].
A Study of full genome of species Streptomyces hygroscopicus var. jinganggensis revealed that a 45kb region of the S. hygroscopicus 5008 chromosome was responsible for the production of secondary metabolite validamycin [1]. Here in, in an attempt to answer the probabilistic enzymatic machinery working behind validamycin pathway, we analyzed the whole cluster reported responsible for the production of validamycin using in silico approach. The interpretation of information about domains and the biochemical reaction steps, for which they could be working, helped to acquire an in-depth understanding of the pathway mechanism. Utilizing this knowledge of protein domain regions within the enzymes improved our filtering criterion of the suitable enzyme among the pool of other enzymes present in the gene cluster. Validation of the methodology was done by comparing the results obtained by in silico approach to those reported in literature for the annotated steps. The analysis revealed some insights about the probable enzymes working at each biochemical step. This work would help in understanding the application of bioinformatics tools and techniques along with understanding the role of fold and domain analysis for revealing the hidden pathways and also to annotate the existing ones. However, this also opens new options for many wet lab experimentations, to prove and explore the probable pathway suggested as a result of this study.
Methodology
Collection of sequence data and gene annotation
The whole genome of S.hygroscopicus and the sequence of the reported gene cluster responsible for validamycin production [2] were taken from the Genbank [15]. Genome annotation of the 45kb gene cluster was done using Genemark S program [16]. Genemark S is a self-training method for prediction of gene- starts in microbial genomes and for finding sequence motifs in regulatory regions. This program was also used to predict the number of genes and related data of the reported gene cluster responsible for validamycin production. The predicted genes were observed for the strands (plus or minus), their lengths, open reading frames they belong to and the protein sequences they code for. To check if the genes cluster contained any rRNA or tRNA forming genes, the RNAmmer 1.2 server [17] and the tRNAscan SE 1.21 [18] were used. RNAmmer predicts 5s/8s, 16s/18s, and 23s/28s ribosomal RNA in the genome sequences and tRNAscan-SE identifies transfer RNA genes in a DNA sequence, both of these are freely available online.
Homology search and structural annotation
All the identified genes from the cluster were used for homology search using BLASTN program [19]. The function of the top hits reported showing maximum similarity with the query sequence was considered as the probable function of the sequence. Besides this, HMMER [20], an online tool to find the probable function of a gene was also used wherein the amino acid sequence of all the genes of the cluster were used as input to find the probable function of the gene. In HMMER, the protein sequences were searched with respect to UniProtKB database using phmmer as the search mode.
All the Genemark S predicted genes of the cluster were then analyzed for their respective location within the whole genome of Streptomyces hygroscopicus on the basis of alignment obtained using BLAST 2 sequence tool [19].
Biochemical pathway characterization
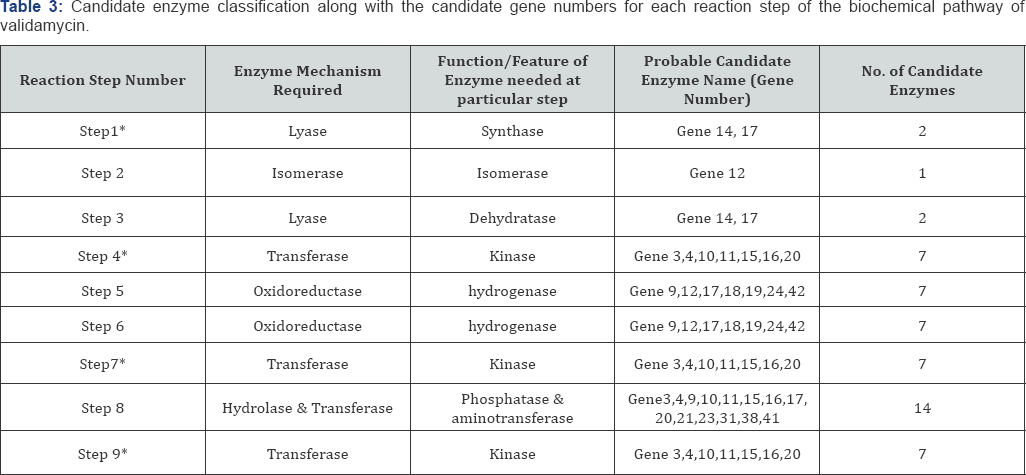
Each step of the proposed biochemical pathway [21] of validamycin production was analyzed for possible metabolic reaction so that the probable enzyme function at each step could be hypothetically deduced. The enzyme found to have the similar function that was required for a particular reaction step to occur was chosen as the probable candidate enzyme for that step and classified under one of the six major enzyme class as per the standard enzyme nomenclature [22]. A hypothesis of enzymatic machinery working behind the pathway was generated and analyzed thoroughly using computational studies.
Computational studies for identification of suitable enzyme from the candidate enzymes
3D structure prediction: The 3D structures prediction of all the genes from the reported cluster was carried out using Phyre2 [23]. Phyre2 is an updated version of Phyre that uses fold recognition technique for predicting the structure and/or function of protein sequence. It is used for detecting remotely homologous structures. It makes profiles (or PSSMs) generated by PSI-Blast for query sequence and the sequences of the known structure. It runs profile-profile matching algorithm together with predicted secondary structure matching. Phyre makes changes to the backbone of the known structure (template) only when modeling of insertions or deletions is needed, by searching its loop library for compatible loops. The similarity as well as query coverage values of the target with respect to its template were analyzed. Procheck module of the SAVS server [24] was used for validation of the generated 3D models using its Ramachandran plot [25] values to check the stereochemical properties of each model. The protein models that were shorter in length were removed.
Binding site identification of the predicted structures: The models were analyzed for their folds and domains using Pfam [26] and SCOP databases [27]. The Pfam database has information related to protein domains and families. It consists of two components: Pfam-A and Pfam-B. Pfam-A consists data of manually curated families. Pfam-B families are automatically generated families which are useful for identifying functionally conserved regions when no Pfam-A entries are found. We can look up the domain organization of a protein model using View a Structure module of the Pfam database. Also we used SCOP database to collect information regarding domains as SCOP (The Structural Classification of Proteins) database contains manual classification of protein structural domains based on their structures and sequences similarities. Collection of available domain regions in the templates of the protein models was done.
Binding site predictions on the basis of volume of void space available along the modeled structure of proteins were done using Sitemap module of Schrodinger [28,29]. Five binding sites were predicted using the Sitemap tool. Structure superposition and sequence alignment of each protein model with its template was manually done to select only those binding sites that lie in the domain region. The sites lying in the domain aligned region, showing similar function identified from the BLASTN [19] homology searches, were used for docking.
Molecular docking studies: All the molecular docking studies were carried out using Schrodinger [28] molecular modeling software. Structures of all the ligands (substrates and products) were built using maestro [30]. The ligands were prepared using LigPrep [31] in which their steroisomeric, tautomeric and ionization states were determined. Various conformations of these prepared ligands were generated using ConfGen [32] module.
The docking studies of all the predicted enzymes were carried out with the substrate of the respective position of the proposed biochemical pathway. The predicted protein structures were prepared using Protein Preparation Wizard in which bond orders were given, hydrogens were added, hydrogen bonds were optimized and energy minimization of the protein structure was done. 3D grid was generated on each identified binding site and flexible docking of the generated conformers was performed onto this grid using Extra Precision scoring function of GLIDE [33,34]. Based on the dock score and hence the binding affinity value of each complex, the most probable enzyme was proposed for carrying out the reaction.
Stability evaluation of the docked complexes: In order to evaluate the stability of the docked complexes and proposing the substrate for the enzymes of the pathway steps, molecular dynamics simulation studies were performed on seven protein- substrate complexes. Desmond 3.1 implemented in Schrodinger 2012 was used for this study [35]. The preparation of the system for each complex was done by solvating protein-substrate complex in simple point charge (SPC) water molecules. The system was neutralized by replacing solvent molecules with the counter ions. Each of the prepared systems was relaxed and equilibrated using minimization and then simulated for 5 nanoseconds (ns) using the multistep protocols devised in Desmond. The minimization was performed by applying a hybrid of steepest descent and the limited memory Broyden- Fletcher-Goldfarb-Shanno (LBFGS) methods with a maximum of 2000 iterations. Two steps of minimization were carried out, first with restraints on solute and second without any restraints. Then a short simulation of 12 picoseconds (ps) was done at 10K temperature with restraints on solute heavy atoms using NVT ensemble (constant number of atoms N, volume V and temperature T). The system was further simulated for 12ps in NPT ensemble at 300K with restraints on heavy solute atoms. The final step of equilibration was performed without any restraints for 24ps using NPT ensemble at 300K. Berendsen thermostats and barostats were used for maintaining the temperature and pressure conditions respectively. Each of the equilibrated systems was simulated for 5ns in NPT ensemble with Nose-Hoover thermostat at 300K and Martyna-Tobias- Klein barostat at 1.01325 bar pressure. The time step of 2 femtoseconds (fs) was used with a recording interval of 1.2ps for energy and 4.8ps for trajectory. The short range electrostatic and vanderwall interactions were truncated at 9.0A and the long- range electrostatic interactions were determined using Smooth particle-mesh Ewald method (PME). Each complex was analyzed for energy plots and RMSD of the protein backbone with respect to the simulation time. The root mean square fluctuations (RMSF) of the backbone were analyzed for each residue.
Results and Discussion
Collection of sequence data
The genome sequence taken from Genbank entry with name Streptomyces hygroscopicus subsp. jinggangensis 5008, complete Genome and having Accession No.: CP003275.1 was found to be 10,145,835 bp long. The sequence was taken from Bioproject No.: PRJNA81087. The source organism was Streptomyces hygroscopicus subsp. jinggangensis 5008. It consisted of full length genome sequence of the S.hygroscopicus subsp. jinggangensis strain 5008.
The gene cluster found to be responsible for producing validamycin was taken from Genbank entry with name Streptomyces hygroscopicus subsp. jinggangensis strain 5008 validamycin cluster, genomic sequence and having Accession No.: DQ164098.1 was found to be of 45164 bp linear DNA length. The source organism was Streptomyces hygroscopicus subsp. jinggangensis 5008. The 45kb sequence contained the 16 structural genes that are reported to run the biosynthetic pathway of validamycin [1].
Gene prediction using Genemark S tool
Initial genome annotation was done using Genemark S program [16], which predicted the genes that were present in the gene cluster. Genemark S analysis was done for the gene cluster sequence which predicted a total of 42 genes as shown in Figure 1. The range of gene lengths varied from 45bps to 2536bps. The class 1 was observed for all predicted 42 genes, showing that all the genes belong to bacterial family. The GC percent of the sequence was reported to be 71.8% in NCBI and as per the Genemark software it was approximately same with 69%.

Each gene of the predicted 42 genes of the cluster was in the tRNA forming genes as was verified using RNAmmer 1.2 [17] coding region and none of these were rRNA forming genes or server and tRNAScanSE server [18] as shown in Figure 1.
Homology searches

Homology searches for each of these 42 genes from the cluster was done using BLASTN program [19] to predict their probable function. The function associated with the top hits considering their percent similarity and coverage length along with significant E value (max. 0.001) were reported as the probable function of the sequence, as shown in Table 1.
Amino acid sequences obtained from the GeneMark S gene prediction tool were used as input to the HMMER online tool to find the probable function and the results obtained were in agreement with the result of BLASTN searches. In HMMER, the protein sequences were searched with respect to UniProtKB database using phmmer as the search mode as shown in Table 1.
Structural annotation
Annotation using GeneMark S tool resulted a list of 42 predicted genes when the input sequence was 45kb long gene cluster sequence. The position of these 42 predicted genes inside the whole genome of Streptomyces hygroscopicus 5008 was found using Blast 2 sequence alignment tool for each gene [19]. The overall region involved was noted in terms of base pair length as shown in Table 1. This gave us an idea about the structural location of the genetic cluster of genes responsible for the validamycin pathway.
Based on the criteria of the alignment, a diagramatic view of the location of the gene cluster was created. This simplified the visualization of the location of the start and end regions of the gene cluster inside the whole genome. The cluster belonged to the region of 355079bps to 399513bps of the whole genome. The gene 1 marked the begining of the sequence cluster at 355079bp followed by genes 2,3,4,5 and 6 in the patch ranging from 355079bp till 361053bp as shown in Figure 1 in region AB. A larger portion B'C of the gene cluster was found in a continous stretch of 361127bp to 391570bp was containing all the 16 reported structural genes. The C'D patch of the Figure 1 streching from 391577bp to 399513bp contained rest of the genes from gene number 33 to gene number 42.
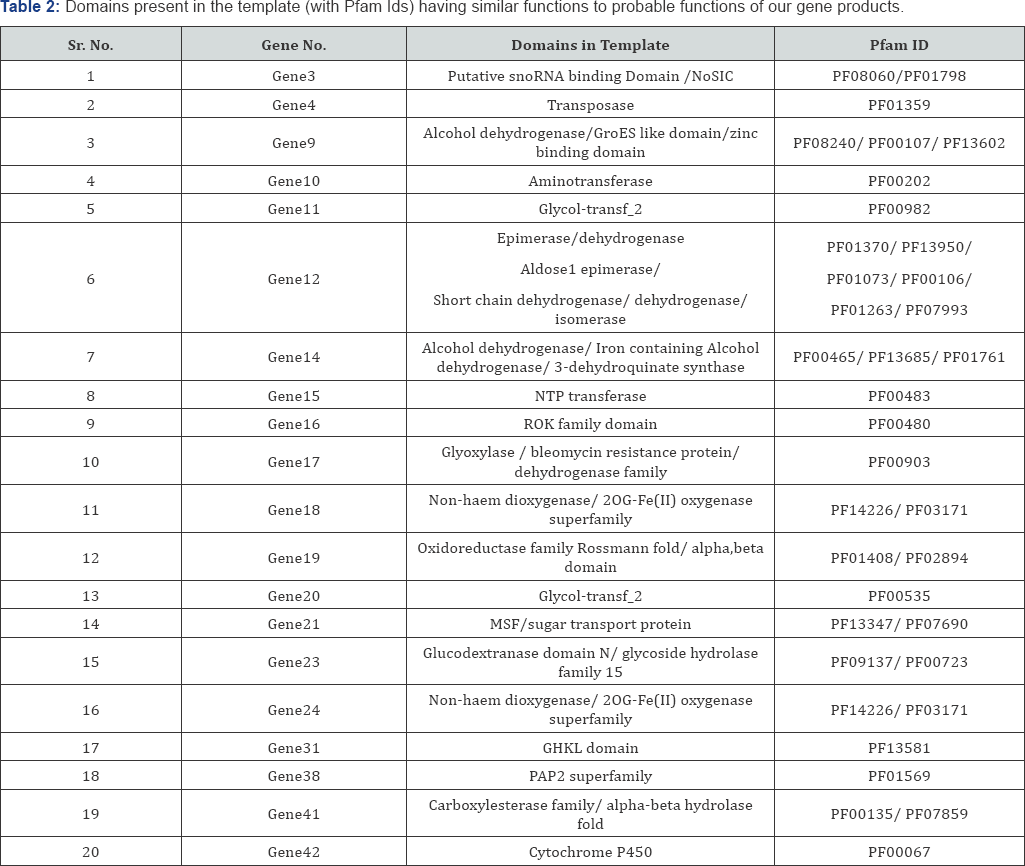
Sixteen structural genes, namely val A to val Q, reported to be responsible for the production of validamycin [1], were aligned with each of the 42 genes of the cluster to identify their particular location, i.e. their corresponding gene number within the 42 predicted genes. Blast 2 sequence tools was used to find the match between the GenBank entry of each val gene with each gene among the 42 predicted genes and the results obtained are discussed in Table 2.
Biochemical pathway characterization
Pathway steps were analyzed and checked for possible metabolic reaction occurring at each step for validamycin biosynthesis so that the probable enzyme working behind it could be hypothetically deduced. There were in all 9 major steps in the pathway as shown in Figure 1. Each reaction of the pathway is discussed in detail (Figure 1) regarding the enzymatic requirement necessary for their catalysis (please refer Supplementary Information).
Candidate enzyme detection/discovery
The results of homology searches helped in elucidating the particular feature of each gene product and that feature was then categorized under the 6 main enzyme classifications. Also the reactions were also generalized based on the same criteria.
However, the proposed pathway steps required enzymatic action belonging to each class of enzyme classification. A total of twelve out of the 42 genes lacked any domain significantly similar to any enzyme class. As nine out of these twelve genes were hypothetical proteins and the rest were having secretory and translocational functions. Thus, these gene products that were not falling under any of the enzyme class required in the pathway reactions were removed from further analysis.
Modeling of probable enzymes
The 3D structures of all the candidate enzymes for each step of the hypothetical pathway were generated. The server used to generate the 3D protein models was Phyre2 and each model was checked for its stability through molecular dynamics simulations of 1ns. The protein model of the gene 16 was not stable as revealed by the stability analysis of the protein structure through the simulation studies. Therefore, the protein sequence of gene 16 was remodeled using Protein Structure Prediction Server (PS2 server) [36]. PS2 is an automated homology modeling server which uses an effective consensus strategy by combining PSI-BLAST, IMPALA, and T-Coffee in both template selection and target-template alignment and it uses MODELLER package to build the final three dimensional structures. The new structure of gene 16 was found to be stable.
The generated models were analyzed with respect to the similarity and query coverage with the template protein used for model building. Also, the Ramachandran plot values for each model were calculated to check the stereochemical properties as discussed in Table 3. Further, ten out of these 30 enzymes were neglected as they were having shorter modeled regions less than 40bps, because of which their binding sites were not generated. Apart from the model length the structure stability (disallowed region of approx. less than 2.6%) was also taken up as a criterion for filtering out the vital enzyme models responsible for the biosynthetic pathway. The ones that were removed were gene numbers 1,2,5,6,8,25,30,32,33 and 34. The remaining 20 candidates were analyzed for domain constituency of their templates.
Domain analysis of templates
The models were analyzed for their folds and domains using databases like Pfam and SCOP. The structure alignment of each model was done with the template to find the position of domains in each model as shown in Table 2. All the domains available in templates were searched using SCOP and Pfam databases and the ones having functionally similarity as required by the modeled enzymes were noted down along with their structural location. Based on the enzyme classification by homology searches and domain based selection criteria, a list of candidate enzymes probably working at each step were tabulated as shown in Table 3.
The region of the modeled proteins having the structural alignment with functionally similar domains were highlighted and manually visualized to find the effective binding site. Each model was then evaluated for their probable binding sites using the Sitemap tool of Schrodinger software. Five to six binding sites were predicted for all these 20 models. The sites lying in the domain aligned region, showing similar function identified from the BLASTN homology searches, were used for docking.
In silico approach to support the results
The docking study for protein-substrate affinity of each enzyme under consideration was done using GLIDE-Ligand docking and their binding affinities were analyzed based on the obtained docking scores. The resultant docking score of enzyme, found to have minimal binding energy values after docking, hinted towards maximum possibility of occurrence of that enzyme than others. This helped in proposing the final biochemical pathway of validamycin.

Analyzing the reaction step 1 in detail: Based on the binding score, as shown in Table 4, obtained by docking the two candidate enzymes namely gene 14 and gene 17, the probable enzyme working as Lyase in step 1 is product of gene 14. The XP GScore of gene14 enzyme product for the substrate is better as compared to the gene 17 enzyme product, which is in agreement with the already annotated result for the step [4]. The ligand interaction of gene14 and D-sedoheptulose-7-phosphate is shown in the Figure 1 and docking image is shown in Figure 1. The OH groups are interacting with various residues like ASP,LYS, ILE and ARG. A total of eight interactions of hydrogen bonds (Table 4) are occurring between the ligand and the receptor.
Analyzing the reaction step 2 in detail: The activity required for the step to occur was that of an isomerase enzyme. Among all the 42 gene products i.e. 42 enzymes only gene 12 was having a functional domain matching to an epimerase domain and showed a possible isomerase character. So, we propose that enzyme synthesised by gene 12 is responsible for the step no. 2 of the biochemical pathway of validamycin synthesis.
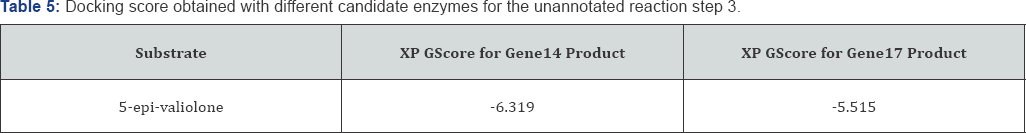
Analyzing the reaction step 3 in detail: The dock scores as shown in Table 5 for the reaction step 3 suggest that the probable enzyme working as Lyase in step 3 is product of gene14 as the XP GScore of gene14 enzyme product for the substrate is better as compared to the gene 17 enzyme product.
The ligand Interaction of gene14 and 5-epi-valilone is shown in the Figure 1 and docking image is shown in Figure 1. The OH groups are interacting with various residues like ASN, LYS, GLU and ASP. A total of six interactions of hydrogen bonds (Table 5) are occurring between the ligand and the receptor.
Analyzing the reaction step 4 in detail: The dock scores obtained for each candidate enzyme for reaction step 4 are shown in Table 6. The binding affinity analysis suggests that the probable enzyme working as transferase in step 4 is product of gene 16. Since the XP GScore of gene16 product for the substrate of the reaction is high as compared to the other candidate enzymes, so enzyme of gene 16 is the enzyme working behind step number 4 which is in agreement with the already annotated data [36,37]. The ligand interaction of gene16 and valienone is shown in the Figure 1 and docking image is shown in Figure 1. The OH groups are interacting with various residues like ASN, ALA, ILE and GLY. A total of six interactions of hydrogen bonds (Table 6) are occurring between the ligand and the receptor.

Analyzing the reaction step 5 in detail: The dock scores as shown in Table 7 for the reaction step 5 suggest that the probable enzyme working as oxidoreductase in step 5 is enzyme product of gene 12. Since the XP GScore of gene 12 for the substrate is high as compared to other candidate enzymes, so gene 12 becomes the proposed enzyme for step number 5. The ligand Interaction of gene12 and valienone-7-phosphate and docking image is shown in Figure 1. The OH groups are interacting with various residues like ALA, LEU, SER and GLY. A total of six interactions of hydrogen bonds (Table 7) are occurring between the ligand and the receptor.

Analyzing the reaction step 6 in detail: The dock scores generated for each candidate enzyme for step 6 as shown in Table 8 suggest that the probable enzyme working as oxidoreductase in step 6 is enzyme product of gene 19. Since the XP GScore of gene 19 for the substrate is high as compared to other candidate enzymes, so gene 19 becomes the proposed enzyme for step number 6. The ligand Interaction of gene19 and valienone is shown in the Figure 1 and docking image is shown in Figure 1. The OH groups are interacting with various residues like ALA, LYS and GLY. A total of three interactions of hydrogen bonds (Table 8) are occurring between the ligand and the receptor.

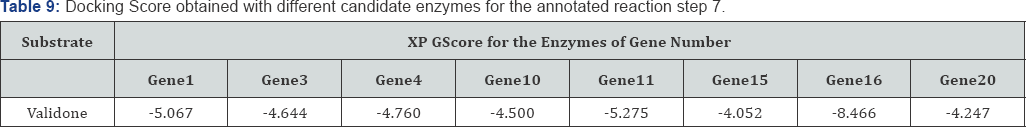
Analyzing the reaction step 7 in detail: Based on the dock score data generated as shown in Table 9 for the XP docking of each candidate enzymes with validone as ligand it could be suggested that the probable enzyme working as transferase in step 7 is product of gene 16. Since the XP GScore of gene16 product for the substrate of the reaction is high as compared to the other candidate enzymes and comparable to gene 11, so enzyme of gene 16 is the enzyme working behind step number 6 which is in agreement with the already annotated data [37]. The ligand Interaction of gene16 and validone is shown in the Figure 1 and docking image is shown in Figure 1. The OH groups of the ligand are interacting with various residues like ASN, ILE and GLU (Table 9).
Analyzing the reaction step 8 in detail: It was seen that the step required a phosphatase as well as aminotransferase activities to catalyze the reaction. The aminotransferase was only enzyme 10 as per the homology searches and domain analysis. Enzyme 31 had BLAST result matching the feature of phosphatase also its domain analysis showed a transferase domain which is an additional advantage as the step requires both the functions of transferase and hydrolase enzyme. Thus, enzyme 10 and enzyme 31 are the proposed enzyme candidates for the step 8 of the validamycin biosynthetic pathway.
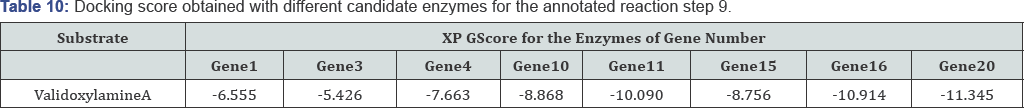
Analyzing the Reaction Step 9 in detail: The dock scores as shown in Table 10 for the reaction step 9 suggest that the probable enzyme working as transferase in step 4 is product of gene 20. Since the XP GScore of gene20 product for the substrate of the reaction is high as compared to the other candidate enzymes, so enzyme of gene 20 is the enzyme working behind step number 6 which is in agreement with the already annotated data [1]. The ligand Interaction of gene20 and validoxylamine A and docking image is shown in Figure 1. The OH groups are interacting with various residues like GLU, THR and ARG. A total of five interactions of hydrogen bonds (Table 10) are occurring between the ligand and the receptor.
The verification of the methodology was done by comparing the results obtained through our in silico approach with the results reported for the annotated steps proven through wet lab experimentations. The results obtained by both methods were in agreement with each other which imply that using this fold based in silico approach for analysis of pathway has driven us to a more concrete platform of obtaining dependable results. The verification suggests that the results obtained for the unannotated steps, using this fold based approach in our in silico analysis of enzymes responsible for the biochemical pathway, would also be reliable.
Molecular dynamics simulation of the docked complexes: Molecular Dynamics (MD) simulations of the seven protein- substrate complexes were carried out. For performing the MD simulation of the gene20 complex in step 9, the domain region of the protein model bound with the substrate was used. The part of the protein between Arg246 and Leu270 was not modeled by the Phyre2 server and analysis of the domain region (using Pfam) of the modeled structure suggested that the domain part has been modeled properly which was lying between Leu270 and Arg420. Sitemap binding site prediction and docking results also suggested that the proposed substrate of the gene20 was binding on the functional domain part of the model and the binding site has no involvement of the other part of the structure (from residues Val8 to Arg246). Therefore the domain region of the gene20 (from Leu270 to Arg420) along with the bound substrate at the binding site was used for carrying MD simulation.
The analysis of the RMSD plots of the protein backbone showed that the protein-substrate complexes of the gene12-step 5, gene14-step 1, gene14-step 3, gene19-step 6 and gene20- step 9 were stable as shown in Figure 1 respectively. Stable trajectories of the RMSD were obtained below 3.5Å for these complexes. MD simulations were performed for the new gene 16 model complexed with the substrates of the reaction step 4 and step 7. These two complexes showed stable trajectories of RMSD below 4Å (Figure 1) that demonstrated the stability of these protein substrate complexes.
Conclusion
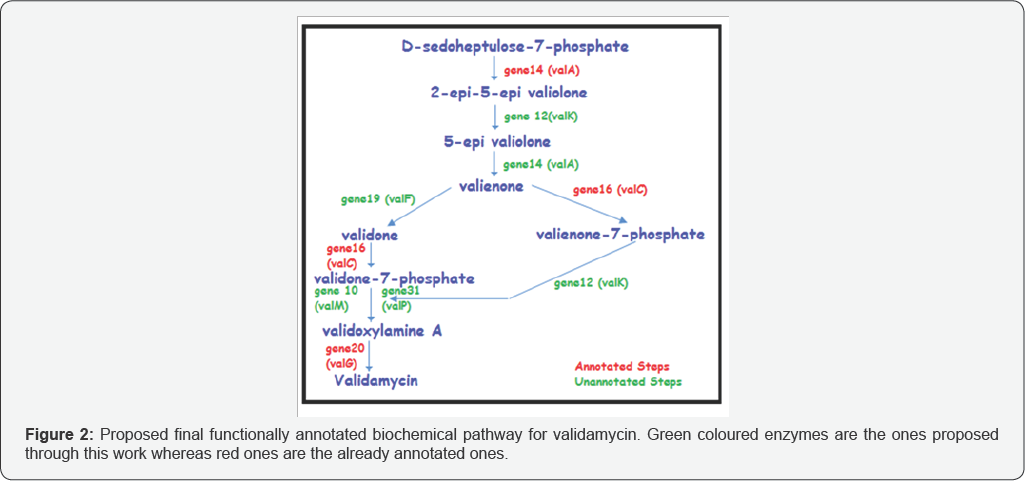
The genes predicted using GeneMark S tool for the reported cluster of Streptomyces hygroscopicus subsp. jinggangensis 5008 were structurally and functionally annotated. Both homology based search and protein fold based selection criteria was adopted to filter out a pool of probable candidate enzymes from the predicted genes for each one of the nine biochemical reaction steps of validamycin pathway, five of which are not annotated till date. The study was aimed to find enzymes involved in the five unannotated pathway reactions of the nine step long validamycin pathway. Homology based searches provided a direct correlation between the enzymatic function and the biochemical changes required for pathway step 8 to occur, thus gene31 (phosphatase) and gene10 (aminotransferase) were hypothesized to be the enzymatic cause of that reaction step. The protein fold based selection helped in finding the possible enzyme i.e. gene12 for the reaction step 2 of the pathway which required an isomerase activity. The remaining three unannotated steps had more than one candidate enzyme, so an in silico based docking approach was used to find the best one among them based on the affinity of enzyme for the substrate. The four previously annotated steps were also analyzed using the same in silico protocol and the results corroborated with the reported in vitro studies for these steps, except for gene16 (valC) in reaction step 7. Further MD simulation studies revealed that all the docked protein- substrate complexes were stable. This study proposes five new enzymes namely gene12, gene14, gene19, gene10 and gene31 for the unannotated steps of validamycin pathway (Figure 2).
Acknowledgment
The authors acknowledge the support provided by the projects BSC0117 and GAP0141.
References
- Bai L, Li L, Xu H, Minagawa K, Yu Y, et al. (2006) Functional analysis of the validamycin biosynthetic gene cluster and engineered production of validoxylamine A. Chem Biol 13(4): 387-397.
- Singh D, Seo MJ, Kwon HJ, Rajkarnikar A, Kim KR, et al. (2006) Genetic localization and heterologous expression of validamycin biosynthetic gene cluster isolated from Streptomyces hygroscopicus var. limoneus KCCM 11405 (IFO 12704). Gene 376(1): 13-23.
- Singh D, Kwon HJ, Rajkarnikar A, Suh JW (2007) Glucoamylase gene, vldI, is linked to validamycin biosynthesis in Streptomyces hygroscopicus var. limoneus, and vldADEFG confers validamycin production in Streptomyces lividans, revealing the role of VldE in glucose attachment. Gene 395: 151-159.
- Yu Y, Bai L, Minagawa K, Jian X, Li L, et al. (2005) Gene cluster responsible for validamycin biosynthesis in Streptomyces hygroscopicus subsp. jinggangensis 5008. Appl Environ Microbiol 71: 5066-5076.
- Zheng L, Zhou X, Zhang H, Ji X, Li L, et al. (2012) Structural and functional analysis of validoxylamine A 7'-phosphate synthase vall involved in validamycin a biosynthesis. PLOS one 7(2): e32033.
- Asano N, Yamaguchi T, Kameda Y, Matsui K (1987) Effect of validamycins on glycohydrolases of Rhizoctonia solani. J Antibiot (Tokyo) 40(4): 526-532.
- Xue YP, Zheng YG, Shen YC (2005) Preparation of trehalase inhibitor validoxylamine A by biocatalyzed hydrolysis of validamycin A with honeybee (Apis cerana Fabr.) BETA-glucosidase. Appl Biochem Biotechnol 127(3): 157-171.
- Li H, Su H, Kim SB, Chang YK, Hong SK, et al. (2011) Enhanced production of trehalose in Escherichia coli by homologous expression of otsBA in the presence of the trehalase inhibitor, validamycin A, at high osmolarity. J Biosci Bioeng 113(2): 224-232.
- Mahmud T, Flatt PM, Wu X (2007) Biosynthesis of unusual aminocyclitol-containing natural products. J Nat Prod 70(8): 13841391.
- Dong H, Mahmud T, Tornus I, Lee S, Floss HG (2001) Biosynthesis of the validamycins: identification of intermediates in the biosynthesis of validamycin A by Streptomyces hygroscopicus var. limoneus. J Am Chem Soc 28: 2733-2742.
- Naganawa H, Hashizume H, Kubota Y, Sawa R, Takahashi Y, et al. (2002) Biosynthesis of the cyclitol moiety of pyralomicin 1a in Nonomuraea spiralis MI178-34F18. J Antibiot 55: 578-584.
- Marina C (2002) Bioinformatics: Bringing it all together technology features. Nature 419: 751-757.
- Edwards JS, Ibarra RU, Palsson BO (2001) In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat Biotechnol 19(2): 125-130.
- Graaf DC, Vermeulen NP, Feenstra KA (2005) Cytochrome P450 in silico: An integrative modeling approach. J Med Chem 48(8): 2725-2755.
- Benson DA, Cavanaugh M, Clark K, Mizrachi IK, Lipman DJ, et al. (2013) GenBank. Nucleic Acids Res 41: D36-D42.
- Besemer J, Alexandre L, Borodovsky M (2001) GeneMarkS: a selftraining method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29(12): 2607-2618.
- Lagesen K, Hallin PF, R0dland E, Sterfeldt HH, Rognes T, et al. (2007) RNammer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res 35(9): 3100-3108.
- Lowe TM, Eddy SR (2005) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5): 955-964.
- Mount DW (2004) Using the basic local alignment search tool (BLAST), Cold spring harbor laboratory press, USA.
- Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39: W29-W37.
- Karp PD, Riley M, Paley SM, Toole PA (2002) The MetaCyc database. Nucleic Acids Res 30(1): 59-61.
- Moss GP (1992) Enzyme nomenclature, world wide web updated version of 1992, School of biological and chemical sciences, Queen mary university of london, Mile end road, London, UK.
- Kelley LA, Sternberg MJE (2009) Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc 4(3): 363-371.
- (2012) SAVS: Version.
- Sheik SS, Sundararajan P, Hussain AS, Sekar K (2002) Ramachandran plot on the web. Bioinformatics 18(11): 1548-1549.
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290-D301.
- Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247(4): 536-540.
- (2012) Schrodinger, Version.
- (2012), Site Map, version 2.6, Schrodinger, New York, USA.
- (2012), Maestro, Version 9.3, Schrodinger, New York, USA.
- (2012), LigPrep Version 2.5, Schrodinger, New York, USA.
- (2012) ConfGen Version 2.3, Schrodinger, New York, USA.
- Friesner RA, Banks Jl, Murphy RB, Halgren TA, Klicic JJ, et al. (2004) GLIDE: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47: 17391749.
- (2012) GLIDE Version 5.8, Schrodinger, New York, USA.
- (2012) Desmond molecular dynamics system, version 3.1, DE Shaw Research, New York, USA.
- Chen CC, Hwang JK, Yang JM (2006) (PS) 2: Protein structure prediction server. Nucleic Acids Res 34: W152-157.
- Minagawa K, Zhang Y, Ito T, Bai L, Deng Z, et al. (2007) A new type of C7-Cyclitol kinase involved in the biosynthesis of the antifungal agent validamycin A. Chembiochem 8(6): 632-641.






























