Selection of Autochthonous Bacterial Starters to Produce Typical Italian Dry-Fermented Sausages with Low Biogenic Amine Content
Simona Guerrini1,2, Manuel Venturi2, Silvia Mangani2, Eleonora Mari1, Lisa Granchi1*and Massimo Vincenzini1
1Department of Food and Microbiological Sciences, University of Florence, Italy
2Food MicroTeam S r l Academic Spin-Off, University of Florence, ItalySubmission: February 02, 2017; Published: June 02, 2017
*Corresponding author: Lisa Granchi, Department of Food and Microbiological Sciences, University of Florence, Italy, Tel:+39-055-2755-916; Email :lisa.granchi@unifi.it
How to cite this article: CSimona G, Manuel V, Silvia M, Eleonora M, Lisa G et. al . Selection of Autochthonous Bacterial Starters to Produce Typical Italian Dry-Fermented Sausages with Low Biogenic Amine Content. Adv Biotech & Micro. 2017; 3(4): 555619. DOI: 10.19080/AIBM.2017.03.555619
Abstract
Development of new lines in wheat and its evaluation under different planting dates is pre requisites for enhancing productivity on sustainable basis. An experiment was conducted at Cereal Crops Research Institute, Pirsabaq, Now shera Pakistan during 2013-14. The study was carried out to evaluate six wheat advanced lines (PR-103, PR-105, PR-106, PR-107, PR-108 and PR-109) on different planting dates (Oct. 25th, Nov. 5th, Nov. 15th, Nov. 25th, Dec. 5th and Dec.15th) to identify the most suitable planting time and judge their performance under late sowing in the central agro-ecological zone of Khyber Pukhtunkhwa (KP), Pakistan. The experiment was laid out in randomized complete block design with split-plot arrangement replicated thrice. Sowing dates effect was studied in main-plots and wheat advance lines in sub-plots. Data were recorded on days to heading, days to maturity, canopy temperature, chlorophyll content, plant height, spike length, spike lets spike-1, grains spike-1, lodging score (%), 1000-kernel weight, grain yield, biological yield and harvest index. Highly significant differences were found among planting dates. Days to heading, days to maturity, canopy temperature, chlorophyll content, plant height and spike length of various wheat advance lines were decreased to 17.6, 19, 11.1, 11.5 15 and 5% respectively when the sowing time was delayed from Oct. 25th to Dec. 15th. Spikelets spike-1, grains spike-1, 1000-grain weight, biomass yield, grain yield and harvest index of various wheat advance lines were also decreased to 7.5, 16, 26.7, 47.7, 35.2 and 19.2% respectively when sowing was delayed from Oct. 25th to Dec. 15th, while lodging score% were maximum on Nov. 5th and zero on Dec. 5th. Grain yield losses of 3.6, 9.9, 12.4, 19.4 and 35.2% were recorded on respective sowing dates and the percent yield losses were increased as sowing delayed from Oct. 25th till Dec. 15th. Significant differences were found among various wheat advance lines on days to heading, spike length, spike lets spike-1, grains spike-1, plant height, lodging score (%), 1000-kernel weight, biological yield while flag leaf area, days to maturity, canopy temperature, and chlorophyll content were not significantly affected. The interactive effects among various wheat advance lines and different sowing dates were not significant except spike lets spike-1. Among various wheat advance lines PR-107 produced higher grain yield (4803kg ha-1) followed by PR-106 and PR-103 while minimum grain yield (3847kg ha-1) were observed for PR-109 at all sowing dates. Among various sowing dates, Oct. 25th sown crop resulted statistically maximum grain yield (5196kg ha-1) followed by Nov. 05th sown crop (5008kg ha- 1) against minimum grain yield (3366kg ha-1) of Dec. 15th sown crops. Average performance of wheat advance lines PR-107 and PR-103 on Oct. 25th and Nov. 5th sowing dates, were recorded (5537kg ha-1) and (5287kg ha-1) respectively. Greater average yield of PR-106 (5373kg ha-1) were recorded on Nov. 15th and Nov. 25th sowing dates. Thus, these results indicates that wheat advance lines should be sown on optimum sowing time Oct 25th and Nov. 05th for producing maximum grain yield in the central agro-ecological zone of Khyber Pukhtunkhwa (KP).
Keywords: Biogenic amines; Lactic acid bacteria; Dry fermented sausages; Starter selection; Autochthonous starter
Introduction
Biogenic amines (BA) are organic bases with aliphatic, aromatic or heterocyclic structures that can be found in several foods [1,2]. These low molecular weight compounds, mainly originating from microbial decarboxylation of precursor amino acids [2-4], possess bioactive properties potentially dangerous to human health [5]. Excessive ingestion of BA can determine action on nervous, gastric and intestinal systems and blood pressure [1]. The most notorious food borne intoxications caused by BA are related to histamine (dilatation of peripheral blood vessels, capillaries and arteries, hypotension, flushing, headache, abdominal cramps, diarrhoea and vomiting) and tyramine (peripheral vasoconstriction, cardiac output increasing, releasing of noradrenaline from the sympathetic nervous system) [2,3]. Nevertheless, it must be taken into account that BA such as putrescine and cadaverine increase the toxicity of histamine and tyramine and can react with nitrite to form heterocyclic carcinogenic nitrosamines, nitrosopyrrolidine and nitrosopiperidine [3]. In humans, under normal conditions, the histamine and tyramine originating from foods are rapidly detoxified by the action of amine oxidases, but in the case of allergic individuals, BA accumulate in the body because of the presence of monoamine oxidase inhibitors [3]. Therefore, the toxicological level of BA is very difficult to be established.
Due to their high protein and amino acid content, dry fermented sausages can be a source of BA [1,2]. In general, tyramine, putrescine, cadaverine and histamine are the most important BA detected in fermented sausages of both industrial and artisan origin [6,7]. However, a high BA accumulation occurs more frequently in dry fermented sausages manufactured in a traditional way, without using starter cultures [2]. These fermented sausages rely on natural contamination by environmental microbiota [8], which may be responsible for distinctive organoleptic qualities [9] that are especially required in typical products. Nevertheless, the indigenous microbial populations carrying out the fermentation are not able to guarantee the safety of artisan-fermented sausages [10] which concerns not only the absence of pathogenic microbiota, but also the presence of BA at low level. The hygienic quality of both raw materials and processing plant is a key factor in BA accumulation during sausage fermentations [11], since most contaminant bacteria, such as enterobacteria and Pseudomonas spp., are known to possess aminogenic capability [1,12-15]. In addition, since the fermentative microbiota, mainly constituted by lactic acid bacteria (LAB), may contribute to BA accumulation during sausage production [1,12-15] in the last years there is an increasing interest in exploiting technological measures to reduce aminogenesis during the manufacture of artisan fermented sausages [14,16,17]. Currently, the use of autochthonous strains unable to produce BA, as starter cultures, seems to be the most effective strategy [2,5,14,18-20]. Indeed, these strains, being well adapted to the specific fermentative process, are very competitive and then potentially able to reduce the activity of indigenous microbiota with aminogenic capability and maintain the distinctive properties of artisan products [5,8,19,21,22]. Therefore, autochthonous strains unable to produce BA could represent a suitable tool in Italian artisan fermented sausages manufacture, since they are still widely produced without using commercial microbial starters [23], although the use of these cultures is known to promote food safety and manufacture standardization [24]. In this perspective, two artisan-fermented sausages obtained from Cinta Senese and Nero Siciliano, two Italian native pig breeds [25], were here considered. These kind of traditional fermented products are in high demand by the Italian consumers [25], but their microbial qualities as well as the manufacturing process are scarcely investigated. Indeed, only two surveys were carried out on Nero Siciliano sausages [26,27], while no investigation was performed on Cinta Senese sausages.
Therefore, after a preliminary investigation on both microbiota and BA concentration of artisan-fermented sausages made with Cinta Senese and Nero Siciliano meats and furnished by different Italian local producers when they were ready for the market, the present work was aimed to select autochthonous LAB starters able to avoid excessive BA accumulation in such fermented sausages.
Material and Methods
Fermented sausages and sampling procedures
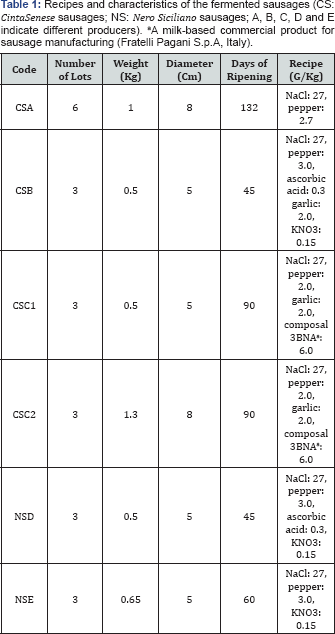
The fermented sausages were traditionally manufactured, according to the recipes reported in Table 1, without using commercial starters by five local producers: three located in Tuscany (A, B and C, producing Cinta Senese sausages coded as CSA, CSB, and CSC) and two in Sicily (D and E, producing Nero Siciliano sausages coded as NSD and NSE). The basic raw materials utilized for sausage manufacturing originated exclusively from the pig breeds taken into consideration, lean pork meat and pork fat being in the range 67-73% and 33-27%, respectively. At the end of the ripening phase, when the products were ready for the market, two whole sausages were taken as representative of each lot and transported to the laboratory under refrigerated conditions (4°C). Totally, 15 CS and 6 NS sausages belonging to different lots produced in the same year were analysed.
Microbiological analysis of the fermented sausages
Ten grams of each sausage were transferred into 90mL sterile physiological solution and homogenized for 5min in a Stomacher Lab Blender 400 (Seward Ltd, Worthing, West Sussex, UK). After decimal dilutions, 100|iL of these suspensions were plated for cell enumeration into specific media [12,28,5,29,30] which were prepared, incubated and used according to the manufacturer's instructions. LAB were counted on MRS agar (Oxoid Ltd, Basingstoke, Hampshire, UK) at pH 5.4 after incubation for 4-5 days at 30°C under anaerobic conditions. Enterococcus spp. was grown on kanamycin aesculinazide agar base (Oxoid Ltd, Basingstoke, Hampshire, UK) at 37°C for 1-2 days. Catalase-positive cocci (CPC) (Staphylococcaceae and Micrococcaceae) were determined on mannitol salt agar (Oxoid Ltd, Basingstoke, Hampshire, UK) after incubation at 30°C for 3 days. Enterobacteriaceae were detected on violet red bile agar (VRBA) with MUG (Oxoid Ltd, Basingstoke, Hampshire, UK) at 37°C for 1 day; Pseudomonadaceae were determined on Pseudomonas agar base, supplemented with glycerol 1mL/100mL and with Pseudomonas CFC supplement (Oxoid Ltd, Basingstoke, Hampshire, UK), after incubation at 25°C for 1-2 days. The technique of spread plate was used for all media with the exception of violet red bile agar, which was inoculated using the pour plate method. For this medium, an overlay was also additionally used as described by APHA (1992) [31].
Determination of biogenic amines in fermented sausages
BA were extracted with trichloracetic acid (0.5g/L) from homogenized samples and determined by Reversed Phase (RP)- HPLC of their dansyl derivatives, according to Marce et al. [32]. A Varian Pro Star Liquid Chromatograph (Varian InsTruments, Inc.), equipped with a fluorimetric detector (Jasco 821-FP), was used. The quantification was carried out with the method of internal standard (heptyl amine) to improve the precision of quantitative analysis. BA separation was performed on a Beckman Ultrasphere ODS 5|im column (250x4.6mm i.d.), according to Lasekan & Lasekan [33]. All analytical data are the mean of two separate determinations.
Lactic acid bacteria identification
About 420 isolates from all MRS plates (about 20 isolates from each lot, all showing Gram positive and catalase negative reactions) were purified and then identified by Amplified Ribosomal DNA Restriction Analysis (ARDRA). DNA was extracted by the method of Reguant and Bordons (2003) [34] and the gene codifying 16Sr DNA was amplified in a thermo cycler (Techne LTD, Cambridge, UK) using the primers FD1 (5-CAACAGAGTTTGATCCTGGCTCAG-3) and RD1 (5-GCTTAAGGAGGTGATCCAGCC-3) described by Weisburg et al. [35]. The PCR reaction mixture, the amplification conditions and the digestion reactions of the amplicons with BfaI, Tru1l and HinfI enzymes (FermentasInc, Burlington, Ontario, Canada) were performed as described by Rodas et al. [36]. The restriction fragments were separated (at 100 volt for 2.5 h) on 0.2g/L agarose gel (Lonza Group Ltd, Basel, Switzerland), containing ethidium bromide (Sigma-Aldrich, St Louis, Missouri, USA) and TBE buffer (1M Tris, 10mM EDTA, 0.9 M boric acid, pH 8.3). The resulting profiles were compared with those reported in the literature [36] and with the profiles of the type strains of some LAB species often found in sausages (Lactobacillus brevis DSM20054, Lactobacillus plantarum DSM20174, Lactobacillus sakei DSM20017). Finally, representative isolates of each ARDRA pattern group were randomly chosen for sequence analysis in order to confirm their identification. Hence, the 16S rDNA PCR amplicons of these isolates were purified using Nucleo Spin Extract II (Macherey-Nagel GmbH & Co. KG, Duren, Germany) and sent to BMR Genomics (Padua, Italy) for sequencing. The sequences obtained in FASTA format were compared with those deposited in Gen Bank DNA database using the basic BLAST search tools.
Intraspecific biodiversity of Lactobacillus sakei
Intraspecific biodiversity of 77 Lactobacillus sakei isolates (51 isolates from the 3 CSC1lots, 22 isolates from NSD, 4 isolates belonging to the GESAAF Department collection and originally isolated from Italian fermented sausages (produced without starter cultures and containing BA concentrations lower than 50 mg/Kg) was analyzed by Randomly Amplified Polymorphic DNA (RAPD), using the following primers: OPL-05 (5-ACGCAGGCA-3), P1 (5-ACGCGCCCT-3), P4 (5-CCGCAGCGTT-3), and MV1 (5-GGACGCTTCTG-3), as reported by Venturi et al. [37]. The random primers MV1 and P4 were used separately, while OPL-5 and P1 were used along with RD1 primer (described above). The DNA amplification was performed as described by Reguant & Bordons [34]. Amplicons were analysed on 1.4% (w/v) agarose gel (Lonza) stained with ethidium bromide (Sigma- Aldrich, St Louis, Missouri, USA) in TBE buffer for 4 h at 80 V and observed by UV transillumination. Gel images were captured as TIFF format files with a CCD camera (UVItec Gel Documentation System, Cambridge, UK). The reproducibility of RAPD-PCR patterns was assessed by comparing the PCR products obtained with different primers and DNA prepared from two separate cultures of the same strains. By combination of the four RAPD profiles for each isolate, according to Venturi et al. [37], a unique dendrogram was obtained by pair-wise comparison of all profiles. Pattern evaluation was made using Dice algorithm and cluster analysis was carried out by the Unweighted Pair Group Method using Arithmetic Averages (UPGMA). Analysis was performed by using Gel Compare 4.0 Software (Applied Maths NV, St-Martens-Latem, Belgium). The calculated cophenetic correlation value for the RAPD dendrogram was 82%, indicating a good level of reliability. The reproducibility between different RAPD-PCR patterns for the same isolate was higher than 82%.
Production of biogenic amines by Lactobacillus sakei isolates
Lactobacillus sakei isolates were cultured under optimal growth condition in their specific culture media grown in MRS broth at 30°C. Once stationary phase was reached, 5mL of each culture were centrifuged (3500xg for 20min), and the cell pellet was resuspended in 5mL of phosphate buffer (0.05M, at pH 6.0) supplemented or not with 100mg/L of each BA precursor (histidine, tyrosine, ornithine and lysine). After 24h of incubation at 30°C, the BA concentration in the buffered cell suspensions was determined by RP-HPLC as above reported.
Technological characterization of the Lactobacillus sakei strains
Fourteen Lactobacillus sakei strains, also including the strain Lactobacillus sakei LM303 isolated from a commercial starter (Textel LM30, Danisco), were grown in MRS broth at 30°C for 24 hours. Once the stationary phase was reached, each culture was streaked on solid media to determine the tolerance of the strains to salts (NaCl, nitrate and nitrite) and to determine the proteolytic and lipolytic activities.
Tolerance to NaCl was determined by colony formation on MRS agar plates, added with different salt concentrations (0.5, 0.7, 1g/L), after 3 days of incubation at 30°C. Likewise, growth capability at different nitrate or nitrite concentrations was determined by colony formation after 3 days of incubation at 30°C on MRS agar plates integrated with NaNO3 (0.2 and 0.3g/L) or with NaNO2 (0.1 and 0.2g/L). The proteolytic activity and the lipolytic activity were tested in Calcium Caseinate and Tributyrin Agar plates (Sigma-Aldrich Chemie GmbH, Switzerland), respectively. Each strain was inoculated on a spot at the surface of the media and, after incubation at 30°C for 3 days, the lipolytic and the proteolytic activity were determined by the appearance of a clear halo surrounding the spots which diameter was measured in mm. Indeed, proteolytic and lipolytic microorganisms metabolize the proteins or lipids present in the media and halos around the colonies appear.
An aliquot of each Lactobacillus sakei culture was also centrifuged (3500xg for 20min) and the cell pellet was then inoculated in liquid media at a concentration of 107CFU/mL to determine the growth capability of the strains at different pH values and under growth conditions simulating some technological parameters of sausage manufacturing.
Growth kinetics at different pH values (5.4, 5.0, 4.5) adjusted by HCl 5N were determined in MRS broth at 30 °C, while growth kinetics and acidification capability under condition simulating technological parameters of sausage manufacturing were determined in SB broth at pH 6.0 and temperature of 15 °C, as reported by Ammor et al. [8]. Bacterial growth on liquid media (MRS at different pH and SB broth) was monitored spectro photometrically at 660nm (spectrophotometer Cary 50 Scan, Varian Inc, Palo Alto, CA, USA). To calculate the kinetic parameters (lag phases, maximum growth rates and growth yields), the growth data were fitted with the Gompertz function and the fit adequacy was checked by the proportion of variance explained by the model (R2) with respect to experimental data (GraphPad Prism 5 software package).
To carry out the selection of Lactobacillus sakei strains based on their technological properties, two virtual isolates, called "the best" and "the worst", were included in the experimental data as reported by Ammor et al. [8]. The virtual results ascribed to "the best" and "the worst" strains were chosenbased on some selection criteria reported in literature [8,38,39]: high growth rate and growth yield, ability to grow at different salt concentrations and at different pH values. Therefore, for each growth condition assayed, the lag phase value assigned to "the best" strain was the minimum value among those recorded for the isolates, while the values for acidification capability as well as for growth rate and growth yield were the maximum ones. To "the best" strain were also attributed virtual positive results from growth under different salt concentrations (assays on plates). Results attributed to "the worst" strain were opposite of those of "the best" one. All the (experimental and virtual) data were statistically treated with cluster analysis as below reported.
Ability assessment of the selected strains to control BA production
To check the correct procedure of selection, two L. sakei strains appropriately chosen were separately inoculated on SB medium, according to the procedure reported above, in presence or not of two BA producer strains. These strains, BL4 and BL6, belonged to Lactobacillus curvatus species, had been previously isolated from Cinta Senese and Nero Siciliano respectively (GESAAF collection). After 8 days of fermentation at 15 °C, chemical analysis to quantify BA, microbial analysis to quantify LAB and molecular analysis to identify LAB were performed as above reported.
Statistical analysis
Microbiological and chemical determinations, performed in duplicate, were elaborated according to one way ANOVA followed by Tukey's test (significance level: p=0.05). Correlation studies between the microbial populations and the individual BA concentration in sausages were carried out by calculating both Pearson and Spearman rank correlation coefficients (significance level: a = 0.05). Cluster analysis of Lactobacillus sakei technological properties was carried out by the Unweighted Pair Group Method using Pearson product-moment correlation coefficient (r). Statistica 7.0 software package was used for all the statistical analysis.
Results
Microbiota and biogenic amine concentrations in Cinta Senese and Nero Sicilian of ermented sausages
The cell concentrations ofthe dominant microbial populations at the end of the ripening phase in different lots of six types of artisan fermented sausages are shown inTable 2. The LAB concentrations ranged between 106 and 109 CFU/g, the lowest values occurring in the CSA sausage characterized by the longest ripening time (132 days). In all the samples, with the exclusion of the CSA1 lot, the cell densities of catalase-positive cocci (CPC) were significantly lower than those of LAB and ranged between values of about 103 and 106 CFU/g. As concerns the spoilage microbiota, the cell densities of both Pseudomonadaceae and Enterobacteriaceae varied between values below the detection limit (<50 and <5 CFU/g, respectively) to values of about 104 CFU/g, but Escherichia coli was never found. A quite similar range of cell concentrations was found for Enterococcus spp., with the exception of the NSD3 lot that showed a higher cell concentration (105 CFU/g).
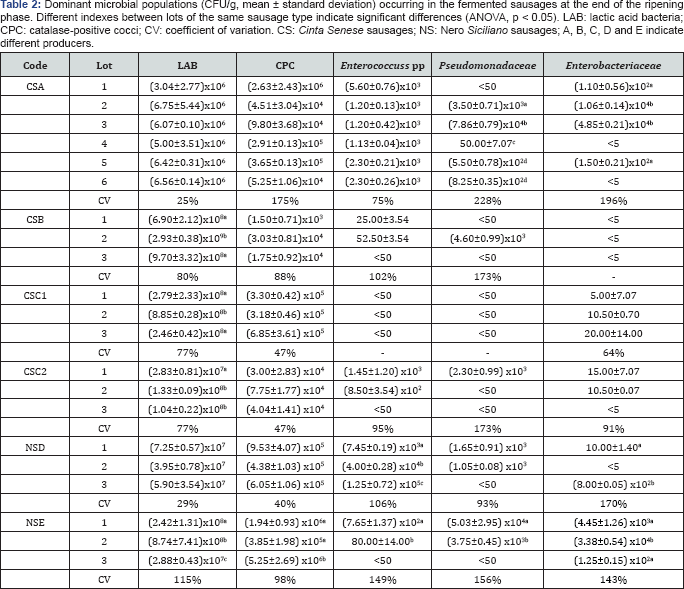
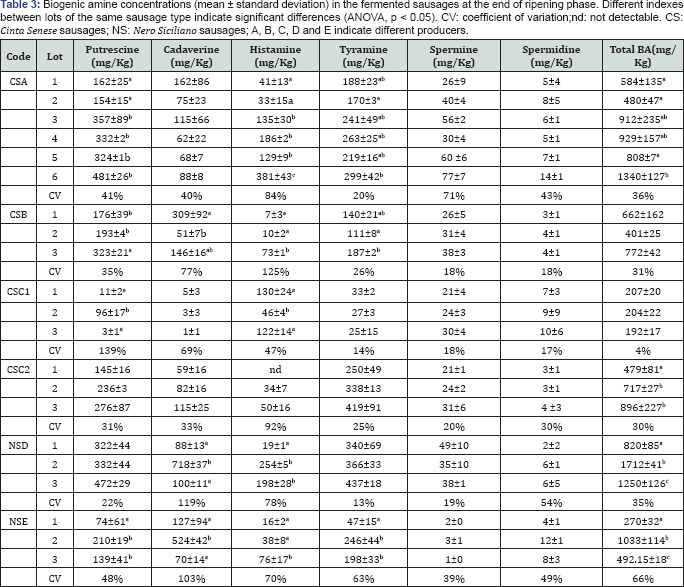
The cell concentrations of the fermentative (LAB and CPC) and/or spoilage microbiota in various lots of the same sausage type showed often differences statistically significant (ANOVA, p<0.05) so that their variation coefficient was often higher than 100%. These observations highlighted the microbial diversity of the artisan fermentative processes. Total BA concentration was found at noticeable values in all the sausage samples, ranging from 191 to more than 1700mg/Kg (Table 3). The variation coefficients of the total BA contents, calculated among different lots of the same sausage type, varied between 4 and 66%. These high variation coefficients as well as the significant differences (ANOVA, p<0.05) in BA contents often observed could be a consequence of the high microbial variability previously described. Finally, correlation analysis between the microbiological data and the concentrations of individual BA in the sausages showed significant positive correlations (α=0.05) only between the cell densities of Enterococcus spp. and the concentrations of both tyramine (Spearman r=0.6065; Pearson r=0.5540) and putrescine (Spearman r=0.5414; Pearson r=0.5560).
The BA concentrations found in many of the assayed sausages pointed out the need to select LAB starter strains potentially able to restrict the activity of indigenous microbiota with aminogenic capability. Therefore, the selection of potential starter strains was carried out among the strains of the LAB species more widespread in the assayed sausages
Identification, characterization and selection of autochthonous LAB strains
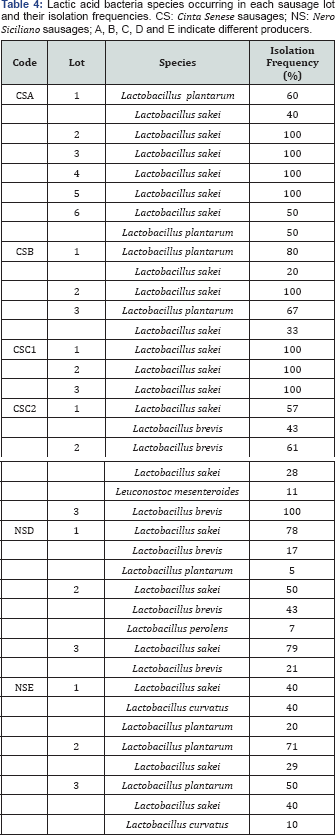
Six different LAB species (Lactobacillus sakei, Lactobacillus plantarum, Lactobacillus brevis, Lactobacillus perolens, Lactobacillus curvatus, Leuconostoc mesenteroides) were found in different lots of the same sausage type, confirming the biodiversity of the fermentative microbiota, which characterized these artisan productions (Table 4). However, Lactobacillus sakei showed the highest isolation frequency in 52% of the assayed lots. Therefore, the selection of potential starters was carried out among the Lactobacillus sakei isolates originated exclusively from sausages showing simultaneously, Lactobacillus sakei as the species with the highest isolation frequency, the lowest cell concentrations of spoilage microbiota as well as the lowest BA concentration. Only the three lots of CSC1 sausage fulfilled the required properties and thus furnished Lactobacillus sakei isolates to be assayed as potential starters for manufacturing Cinta Senese sausages. Since none of the Nero Siciliano sausages fulfilled the required properties, potential starters from this type of sausage were chosen only from the lots showing Lactobacillus sakei as the species with an isolation frequency higher than 70% (Table 4).
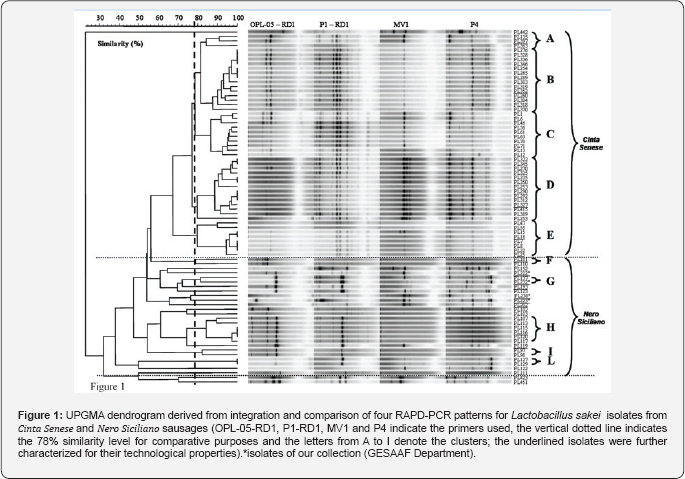
All the chosen isolates (Table 5) and four isolates belonging to the GESAAF Department collection, were typed by molecular methods to characterize them at the strain level and, hence, to perform the planned technological characterization only on different strains or biotypes. Genotypic characterization of the isolates was carried out using a combination of conventional and multiplex RAPD-PCR reactions and the resulting UPGMA dendrogram, shown in Figure 1, describes the similarity relationship among all the isolates. Based on a cut-off value of 78%, 59 out of 77 isolates grouped into 8 clusters. Four clusters (A to D) included all the isolates from Cinta Senese sausages, three clusters (F, H, and I) included all the isolates from Nero Siciliano sausages and one cluster (G) included one isolate from Nero Siciliano (PL137) and one from GESAAF Department collection (PL232). Three isolates from Cinta Senese (PL423, PL451, PL442), eleven from Nero Siciliano (PL202, PL193, PL152, PL123, PL122, PL119, PL111, PL102, PL103,PL127, PL129) and three from GESAAF collection (PL230, PL223, PL222) were well separated each other and from other isolates. Therefore, cluster analysis of RAPD patterns highlighted the presence of different biotypes within Lactobacillus sakei isolates, confirming the high degree of heterogeneity of this LAB species [23]. Nevertheless, results suggested also the presence of manufacture-specific strains of Lactobacillus sakei. Indeed, the assayed Lactobacillus sakei were isolated from sausages produced in two manufactures (CSC and NSD), and according to the dendrogram obtained, clusters from A to D comprised only isolates originating from sausages produced in the CSC manufacture while the remaining clusters included isolates from sausages produced in the other manufactures, with the exception of two strains, PL423 and PL451.
Since starter cultures should be unable to produce BA, the isolates were also assayed for their aminogenic capability. Quantitative analysis demonstrated that more than 40% of the isolates were able to produce histamine, even if at low concentrations (from 2 to 9mg/L after 24 hours in phosphate buffer supplemented with 100mg/L histidine). In this connection, also Freiding et al. [40] demonstrated that some strains of Lactobacillus sakei were able to produce histamine, but with a lower frequency (almost 2% of the tested isolates).
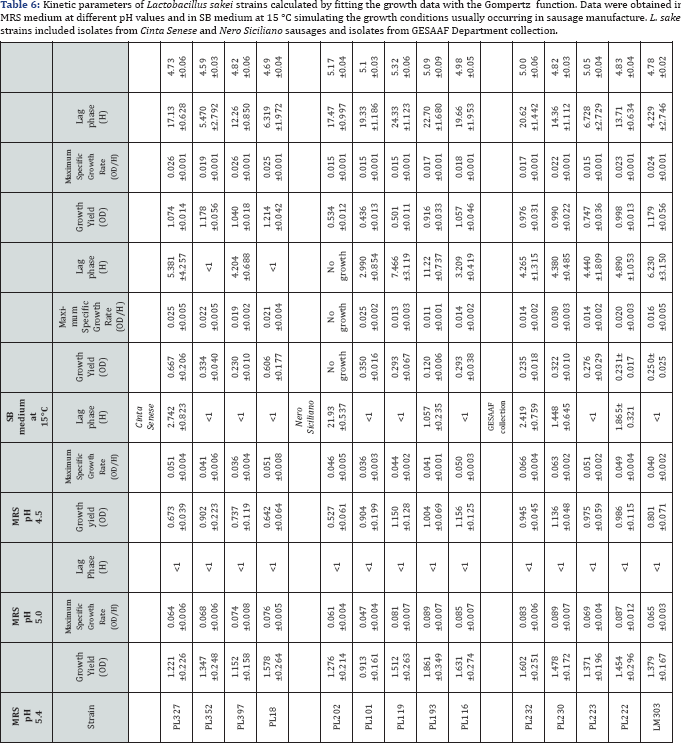
13 Lactobacillus sakei strains (underlined in Figure 1), representative of the various biotypes lacking aminogenic capability, were characterized for their technological performances to detect strains potentially able to reduce aminogenesis by competing with indigenous microbiota. The results from this technological characterization demonstrated that all the strains were able to grow in the presence of 5 and 7g/100mL NaCl as well as in the presence of NaNO2 (0.1 and 0.2g/L) and NaNO3 (0.2 and 0.3g/L), with the exception of the strains PL230 and PL232 that grew poorly in the presence of 0.3 g/L NaNO3. None of the assayed strains was able to grow at 10g/100mL NaCl and to display appreciable proteolytic and lipolytic activities. All the strains were also able to grow at the different pH values, with the exception of the strain PL202 which was unable to grow at pH 4.5 (Table 6). Lag phase was lower than 1 hour in 100% and in 57% of the strains cultured at pH 5.4 and 5.0, respectively. At pH 4.5, only two strains isolated from Cinta Senese sausages showed a lag phase lower than 1 hour, the other strains showing values ranging between 3 and 11 hours (Table 6). In any case, all the strains showed the reduction of their growth performances when pH values decreased. On SB medium, simulating growth conditions as those usually occurring in sausage manufacture at the temperature of 15%C, the best performances were attained by the biotypes PL18 and PL352, both originated, from sausages made with Cinta Senese meat (Table 5).
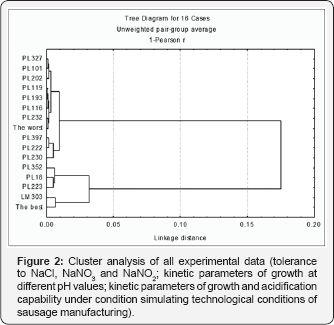
Cluster analysis of all experimental data grouped the strains into two clusters at a distant linkage of 0.17 (Figure 2). One cluster included the virtual strain indicated as "the best", the commercial starter LM303, two strains isolated from Cinta Senese (PL18 and PL352) and one strain belonging to GESAAF Department collection (PL223). All the other 11 strains grouped in the second cluster, together with the virtual strain indicated as "the worst". All the strains isolated from Nero Siciliano were included in this cluster, so that no strain isolated from this type of sausage should be suitable as a starter culture. The presence of the commercial starter strain in the first cluster highlights the reliability of the strain selection procedure here carried out. Hence, PL18 and PL352 could be actually proposed as autochthonous starter strains to be tested in a real manufacture of fermented sausages from Cinta Senese meat.
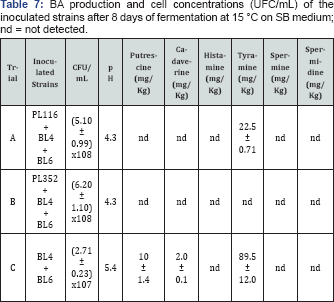
Ability assessment of the selected strains to control BA production
Finally, to further verify the soundness of the strain selection procedure here carried out, one strain belonging to the first group (PL352) and one to the second (PL116) were selected to assess their capabilities to avoid BA accumulation from autochthonous LAB. These two strains were separately inoculated on SB medium in presence of other two strains (BL4 and BL6) belonging to Lactobacillus curvatus species, isolated from sausages and able to produce tyramine, cadaverine and putrescine. To simulate the sausage fermentation with a commercial starter, PL116 and PL352 were inoculated at concentrations of 106UFC/mL, while the BA producer LAB (BL4 and BL6) at concentrations of 104UFC/mL (A and B trials respectively). The BA producer LAB were also inoculated alone to verify their ability to produce putrescine, cadaverine and tyramine (C trial). After 8 days of fermentation, BA concentrations and vital counts were assayed (Table 7). Only PL352 was able to maintain low concentrations of BA when BA producer LAB were present.
Discussion
The BA found in the assayed artisan sausages were histamine, tyramine, putrescine, cadaverine, spermine and spermidine. Correlation studies between the microbiological data and the concentrations of BA in theanalysed sausages seemed to demonstrate that the BA producing capability was widely distributed within the various microbial populations occurring in the fermented sausages, in agreement with other Authors [1,41,42]. Despite the toxicological level of BA is very difficult to be established, in 4 sausage samples the total BA content exceeded the value of 1000mg/Kg that is the limit considered dangerous for health according to Santos [3]. In the same way, the concentrations of histamine (ranging between values below the detection limit to 361mg/Kg) and of tyramine (ranging between 47 to 437mg/Kg), occurring in the assayed sausages were higher than the values reported in literature [1]. Considering the synergistic interaction between histamine and tyramine [43], measures to reduce BA formation in the fermented sausages studied are strongly recommended. Currently, the use as starter cultures of autochthonous strains unable to produce BA seems to be the most effective strategy. In fact, as reported by Latorre-Moratalla et al. [5], the use of a competitive starter culture should prevent the growth of microbiota responsible for BA formation during sausage manufacture. The strategy to use autochthonous starter strains, besides to improve both quality and safety of artisan-fermented sausages, should maintain the typical organoleptic profile of the final products. These aspects could be taken into account, especially in view of a hygienic and organoleptic standardization of artisan fermentative processes. However, the success of this strategy is strictly dependent on the choice of suitable systems to select the potential starter strains. In this work, the selection program to individuate the potential starter strains was characterized by two phases. The first phase was to isolate strains only from sausages simultaneously characterized by a dominant Lactobacillus sakei population, and low densities of spoilage microbiota as well as by low concentrations of BA. Indeed, the Lactobacillus sakei species was chosen to select starter strains because it is considered particularly well adapted to the environment of fermented sausages [8,23,29,44] and more competitive than other lactobacilli [45]. The second phase of the selection program was to select, within the group of bacterial strains detected in the first phase, those potentially able to reduce aminogenesis by competing with indigenous microbiota. A laboratory experiment demonstrated that the selection procedure used in this study was actually effective to select a strain able to reduce aminogenesis by competing with BA producer LAB strains.
In addition, the presence of different biotypes of Lactobacillus sakei in sausages produced in different manufactures could be due to the strong selective effect of the manufacturing conditions on the indigenous microbiota. Consequently, a typical Lactobacillus sakei population, as otherwise suggested also in other studies [23,46,30], characterized each manufacture. This different Lactobacillus sakei strain, that is different biotypes, could possibly contribute to the typical characteristics of the artisanal sausages produced in a specific manufacture besides to assure healthier products.
Conclusion
Accumulation of biogenic amines in artisan fermented sausages obtained with Cinta Senese and Nero Siciliano meat was demonstrated in this study. The use of autochthonous strains unable to produce BA could represent a suitable tool in these fermented sausages manufacture. This work may be considered the proposal of a suitable strategy in developing autochthonous starters for the manufacture of typical fermented sausages. The next stepsshould be to estimate the effective influence of the selected strains on the hygienic and sensory characteristics of the final products in a real industrial production.
References
- Suzzi G, Gardini F (2003) Biogenic amines in dry fermented sausages: a review. Int J Food Microbiol 88(1): 41-54.
- Gardini F, Ozogul Y, Suzzi G, Tabanelli G, Ozogul F (2016) Technological Factors affecting Biogenic Amine Content in Foods: A Review. Front Microbiol 7: 1218.
- Silla Santos MH (1996) Biogenic amines: their importance in foods. Int J Food Microbiol 29(2-3): 213-231.
- ten Brink B, Damink C, Joosten HM, Huis in't Veld JH (1990) Occurrence and formation of biologically active amines in foods. Int J Food Microbiol 11(1): 73-84.
- Latorre-Moratalla ML, Bover-Cid S, Vidal-Carou MC (2010) Technological conditions influence aminogenesis during spontaneous sausages fermentation. Meat Science 85(3): 537-541.
- Miguelez-Arrizado MJ, Bover-Cid S, Latorre-Moratalla ML, Vidal- Carou MC (2006) Biogenic amines in Spanish fermented sausages as a function of diameter and artisanal or industrial origin. J Sci Food Agric 86: 549-557.
- Parente E, Martuscelli M, Gardini F, Grieco S, Crudele MA, et al. (2001) Evolution of microbial populations and biogenic amine production in dry sausages produced in southern Italy. J Appl Microbiol 90(6): 882891.
- Ammor S, Dufour E, Zagorec M, Chaillou S, Chevallier I (2005) Characterization and selection of Lactobacillus sakei strains isolated from traditional dry sausage for their potential use as starter cultures. Food Microbiol 22: 529-538.
- Lebert I, Leroy S, Giammarinaro P, Lebert A, Charcornac JP, et al. (2007) Diversity of microorganisms in the environment and dry fermented sausages of small traditional French processing units. Meat Sci 76(1): 112-122.
- Baka AM, Papavergou EJ, Pragalaki T, Bloukas JG, Kotzekidou P (2011) Effect of selected autochthonous starter cultures on processing and quality characteristics of Greek fermented sausages. LWT-Food Science and Technology 44: 54-61.
- Tasic T, Ikonic P, Mandic A, Jokanovic M, Tomovc V, et al. (2012) Biogenic amines content in traditional dry fermented sausage Petrovskaklobasa as possible indicator of good manufacturing practice. Food Control 23(1): 107-112.
- Bover-Cid S, Hugas M, Izquierdo-Pulido M, Vidal-Carou MC (2001) Amino acid-decarboxylase activity of bacteria isolated from fermented pork sausage. Int J Food Microbiol 66(3): 185-189.
- Ruiz-Capillas C, Jimenez-Colmenero F (2004) Biogenic amines in meat and meat products. Crit Rev Food Sci Nutr 44(7-8): 489-499.
- EFSA Panel on Biological Hazards (BIOHAZ) (2011) Scientific Opinion on risk based control of biogenic amine formation in fermented foods. EFSA J 9(10): 2393.
- Bover-Cid S, Latorre-Moratalla ML, Veciana-Nogues MT, Vidal-Carou, MC (2014) Biogenic amines. In: Motarjemi Y, Moy GG, Todd ECD (Eds.), Encyclopedia of Food Safety, Elsevier Inc, Amsterdam 2: 381-391.
- Latorre-Moratalla ML, Veciana-Nogues T, Bover-Cid S, Garriga M, Aymerich T, et al. (2008) Biogenic amines in traditional fermented sausages produced in selected European countries. Food Chem 107: 912-921.
- Chong Xie, Hu-Hu Wang, Xiao-Kai Nie, Lin Chen, Shao-Lin Deng, Xing-Lian Xu (2015) Reduction of biogenic amine concentration in fermented sausage by selected starter cultures. CyTA - Journal of Food 13(4): 491-497.
- Benito MJ, Martin A, Aranda E, Perez-Nevado F, Ruiz-Moyano S (2007) Characterization and selection of autochthonous lactic acid bacteria isolated from traditional Iberian dry-fermented salchichon and chorizo sausages. J Food Sci 72(6): M193-201.
- Latorre-Moratalla ML, Bover-Cid S, Talon R, Carriga M, Zanardi E, et al. (2010) Strategies to reduce biogenic amines accumulation in traditional sausage manufacturing. LWT- Food Science Technology 43: 20-25.
- Villani F, Casaburi A, Pennacchia C, Filosa L, Russo F, et al. (2007) Microbial ecology of the soppressata of Vallo di Diano, a traditional dry fermented sausage from southern Italy, and in vitro and in situ selection of autochthonous starter cultures. Appl Environ Microbiol 73(17): 5453-5463.
- Callejon S, Sendra R, Ferrer S, Pardo I (2014) Identification of a novel enzymatic activity from lactic acid bacteria able to degrade biogenic amines in wine. Appl Microbiol Biotechnol 98(1): 185-198.
- Guarcello R, De Angelis M, Settanni L, Formiglio S, Gaglio R, et al. (2016) Selection of Amine-Oxidizing Dairy Lactic Acid Bacteria and Identification of the Enzyme and Gene Involved in the Decrease of Biogenic Amines. Appl Environ Microbiol 82: 6870-6880.
- Bonomo MG, Ricciardi A, Zotta T, Parente E, Salzano G (2008) Molecular and technological characterization of lactic acid bacteria from traditional fermented sausages of Basilicata region (Southern Italy). Meat Sci 80(4): 1238-1248.
- Baruzzi F, Matarante A, Caputo L, Morea M (2006) Molecular and physiological characterization of natural microbial communities isolated from a traditional Southern Italian processed sausages. Meat Science72(2): 261-269.
- Pugliese C, Sirtori F (2012) Quality of meat and meat products produced from southern European pig breeds. Meat Science 90(3): 511-518.
- Francesca N, Sannino C, Moschetti G, Settanni L (2013) Microbial characterisation of fermented meat products from the Sicilian swine breed "Suinonerodei Nebrodi”. Annals Microbiol 63(1): 53-62.
- Moretti VM, Madonia G, Diaferia C, Mentastia T, Paleari MA, et al. (2004) Chemical and microbiological parameters and sensory attributes of a typical Sicilian salami ripened in different conditions. Meat Sci 66(4): 845-854.
- Iacumin L, Manzano M, Comi G (2012) Catalase-positive cocci in fermented sausage: Variability due to different pork breeds, breeding systems and sausage production technology. Food Microbiol 29(2): 178-186.
- Papamanoli E, Tzanetakis N, Litopoulou-Tzanetaki E, Kotzekidou P (2003) Characterization of lactic acid bacteria isolated from a Greek dry-fermented sausage in respect of their technological and probiotic properties. Meat Sci 65(2): 859-867.
- Rantsiou K, Cocolin L (2006) New developments in the study of the microbiota of naturally fermented sausages as determined by molecular methods: A review. Int J Food Microbiol 108: 255-267.
- APHA (1992) Compendium of methods for the microbiological examination of foods (3rd edn), American Public Health Association, Washington, DC, USA.
- Marce M, Brown DS, Capell T, Figueras X, Tiburcio AF (1995) Rapid high-performance chromatographic method for the quantization of polyamines as their dansyl derivatives: application to plant and animal tissues. J Chrom 666: 329-335.
- lasekan OO, Lasekan WO (2000) Biogenic amines in traditional alcoholic beverages produced in Nigeria. Food Chem 69: 267-271.
- Reguant C, Bordons A (2003) Typification of Oenococcus oeni strains by multiplex RAPD-PCR and study of population dynamics during malolactic fermentation. J ApplMicrobiol 95:344-353.
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173(2): 697-703.
- Rodas AM, Ferrer S, Pardo I (2003) 16S-ARDRA, a tool for identification of Lactic Acid Bacteria isolated from grape must and wine. System Appl Microbiol 26(3): 412-422.
- Venturi M, Guerrini S, Granchi L, Vincenzini, M (2012) Typing of Lactobacillus sanfranciscensis isolates from traditional sourdoughs by combining conventional and multiplex RAPD-PCR profiles. Int J Food Microbiol 156(2): 122-126.
- Buckenuskes HJ (1993) Selection criteria for lactic acid bacteria to be used as starter cultures for various food commodities. FEMS Microbiol Rev 12: 253-271.
- Hozapfel WH (2002) Appropriate starter culture technologies for small-scale fermentation in developing countries. Int J Food Microbiol 75(3):197-212.
- Freiding S, Gutsche KA, Ehrmann MA, Vogel RF (2011) Genetic screening of Lactobacillus sakei and Lactobacillus curvatus strains for their peptidolytic system and amino acid metabolism, and comparison of their volatilomes in a model system. System Appl Microbiol 34(5): 311-320.
- Guerrini S, Mangani S, Franci O, Vincenzini M (2007) Biogenic amine producing capability of bacterial populations isolated during processing of different types of dry fermented sausages. Italian Journal of Animal Science 6: 688-690.
- Laranjo M, Gomes A, Agulheiro-Santos AC, Potes ME, Cabrita MJ, et al. (2017) Impact of salt reduction on biogenic amines, fatty acids, microbiota, texture and sensory profile in traditional blood dry-cured sausages. Food Chem 218: 129-136.
- Del Rio B, Redruello B , Linares DM, Ladero V, Fernandez M, et al. (2017) The dietary biogenic amines tyramine and histamine show synergistic toxicity towards intestinal cells in culture. Food Chem 218: 249-255.
- Latorre-Moratalla ML, Bover-Cid S, Veciana-Nogues MT, Vidal-Carou MC (2012) Control of biogenic amines in fermented sausages: role of starter cultures. Front Microbiol 3: 169.
- Wang XH, Ren HY, Liu DY, Zhu WY, Wang W (2013) Effects of inoculating Lactobacillus sakei starter cultures on the microbiological quality and nitrite depletion of Chinese fermented sausages. Food Control 32: 591596.
- Cocolin L, Dolci P, Rantsiou K (2011) Biodiversity and dynamics of meat fermentations: The contribution of molecular methods for a better comprehension of a complex ecosystem. Meat Sci 89(3): 296302.






























