The Place and Importance of Pcr-Rflp Method in Determination of Mycobacteria Species in Routine Laboratory Practice
Gülnur Tarhan1*, Tanil Kogagöz2, Salih Cesur3 and İsmail Ceyhan4
1Department of Medical Microbiology, Adiyaman University, Turkey
2Department of Medical Microbiology, Acibadem University, Turkey
3Department of Infectious Diseases, Ankara Training and Research Hospital, Turkey
4Atatürk Chest Diseases and Thoracic Surgery Education and Research Hospital, Turkey
Submission: February 10, 2017; Published: May 04, 2017
*Corresponding author: Gülnur Tarhan, Department of Medical Microbiology, Adiyaman University, Turkey, Email: gulnur.tarhan@yahoo.com
How to cite this article: Gülnur T, Tanil K, Salih C, ysmail C. The Place and Importance of Pcr-Rflp Method in Determination of Mycobacteria Species in Routine Laboratory Practice. Adv Biotech & Micro. 2017; 3(3): 555612.DOI: 10.19080/AIBM.2017.03.555612.
Abstract
Background: Rapid identification of mycobacterial species is the most important step for effective and true treatment. Various molecular methods have been used for rapid identification of mycobacterial species. In this study, we were aim to evaluate the place PCR-RFLP method in routine use and its application practice by the hsp65 gene PCR amplification from clinical isolates.
Material and methods: In our study, 80 mycobacterial isolates and 21 reference ATCC strains were evaluated to by in house PCR -RFLP of an amplified 439-bp segment of the gene encoding the 65-kDa heat shock protein. Restriction digests amplicons were separated by 8 % polyacrylamide and 4% Nusieve agarose gel electrophoresis (PAGE). By including a size standard in each sample, the restriction fragment profile was evaluated using algorithm.
Results: Of 80 mycobacteria strains, 70 (87.5%) were identified as Mycobacterium tuberculosis complex and 4 (5%) as Mycobacterium scrofulaceum, 3 (3.75%) as Mycobacterium gordonae. 3 (3.75%) strains displaying different restriction patterns could not be identified as they were found in the algorithm reported in the literature.
Conclusion: PCR-RFLP method is a rapid, practical and reliable method that can be used routinely in laboratories for typing mycobacterium strains isolated from clinical samples. Restriction enzymes and molecular weight standards play an important part in the efficacy of the test. It is suggested that quality control of materials used should be made comparatively with standards before the test and that more than one molecular weight standards at proper band size should be used in order to minimise mistakes at the stage of evaluation.
Keywords: Mycobacteria; PCR-RFLP; hsp65 gene
Introduction
Tuberculosis (TB) is one of the most ancient infectious diseases of human beings, and it is still in a leading position among infectious diseases as a cause of morbidity and mortality. In recently years, the HIV epidemics and the use of drugs that depress the immun system have resulted in an increase in infections [1,2]. Mycobacteria other than Mycobacterium tuberculosis (MOTT) infections are getting increase in human [3]. Key factors in the control of tuberculosis are rapid detection, adequate therapy, and contact tracing to arrest further transmission [4,5]. Determination of the species of mycobacteria, that are active agents in mycobacteria infections is important in developing appropriate treatment protocols and collecting epidemiological data. Conventional biochemical methods and phenotypic tests for species differentiation of Mycobacteria are laborious and time-consuming and frequently require specialized testing that is beyond the capacity of clinical laboratories. Various genotypic methods for the rapid identification of mycobacteria have been used in recent years [6-9]. With in the these methods, polymerase chain reaction-restriction fragment length polymorphism (PCR- RFLP) method was described by Plikaytis et al. for differentiating among slowly growing Mycobacterium species by PCR and restriction fragment length polymorphism analysis in 1992 [10]. This method is an easy, rapid and inexpensive method of identifying mycobacteria species in a single experiment when comprasion with the other molecular typing methods. This technique has been applied to several genes, for example 16S ribosomal DNA (rDNA), hsp65, dnaJ, groES and rpoB genes. The fundamental mechanism of this method is based on evaluation of the gene coding for the 65-kDa heat shock protein by PCR and restriction enzyme analysis. The PCR-RFLP was routinely used for the taxonomic separation of rapidly growing mycobacteria, identification of mycobacteria strains [11-14].
The success of this method, that yields results quite rapidly, varies according to standardization of the method used of each step, activity of restriction enzymes employed, markers of molecular weight, algorithm and experience of the user. In all these studies the algorithm describing the mycobacteria species is based on the use of two restriction enzymes (BstEII and HaeIII) and separation of the restriction fragments on an agarose gel. PRA patterns are then interpreted by converting the running distance in electrophoresis to apparent molecular size. Difficulties in PCR- RFLP interpretation may stem from similarities in a number of band sizes that are critical for discrimination of species and are not sufficiently resolved by agarose-based gel electrophoresis [15-19]. In this study, we were aim to evaluate the place PCR- RFLP method in routine use and its application practice by the hsp65 gene PCR amplification.
Material and Methods
Bacterial strains
80 mycobacterial isolates and 21 reference ATCC strains (M. bovis BCG ATCC 27291, M. smegmatis ATCC 14468, M. kansasii ATCC 12478, M. fortuitum ATCC 6841, M. parafortuitum ATCC 19686, M. fortuitum subs. fortuitum ATCC 6841, M. abscessus ATCC19977, M. chelonea subs. chelonea ATCC 14472, M. scrofulaceum ATCC 19981, M. szulgai ATCC 35799, M. gordonae ATCC 14470, M. intracellulare ATCC 13950, H37Rv ATCC 25618, H37Ra ATCC 25177, M. xenopi ATCC 19250, M. avium ATCC 25291, M. triviale ATCC 23292, M. diernhoferi ATCC 19340, M. marinum ATCC 927, M. terrae ATCC 15755, M. haemophilum ATCC 29548) were evaluated to by in house PCR- RFLP. Twenty-one reference strains were obtained from Prof. Dr. Tanil Kocagoz, Acibadem Universitiy of Turkey. The mycobacterial isolates were obtained from the clinical samples submitted to National Tuberculosis Reference and Research Laboratory of Refik Saydam Hygiene Center, Ankara, Turkey.
Biochemical tests and growth characteristics
Bacteriologic determination of clinic isolates was base on growth rate, colony morphology, pigmentation, bichemical tests [2-20].
Bacterial strains
A loopful of cells were suspended in 500µl of TE buffer (10mM Tris , 1mM EDTA , pH 8.0 ) in a 1.5ml screw- cap microcentrifuge tube. The suspension was centrifuged at 15,000xg for 10min. The supernatant was removed and then the pellet was washed twice with 500µl of TE buffer and then resuspended with 200µl of the same buffer. The samples were incubated in a boiling water bath for 20 min centrifuged, and supernatants containing DNA was transferred to clean microcentrifuge tubes and kept at - 20°C until used [13].
DNA Amplification
A total of 10µl of the DNA-containing supernatant was added to a reaction tube containing 50µl PCR mixture (1X PCR buffer, 1X PCR Enhancer, 2.5mM MgCl2, 200-µM (each) deoxyribonucleoside triphosphate, 50pM (each) primer and 1.25 U of Taq poymerase [Fermentase]). The reaction tube was first heated for 3min at 94 °C and then to 44 cycles of amplification (1min 94 °C, 1min at 58 °C, 1min 30s at 72 °C); this was followed by 4min of extension at 72 0C. Primers Tb11 (5-ACCAACGATGGTGTGTCCAT) and Tb12 (5- CTTGTCGAACCGCA TACCCT) amplified a 441 bp fragment between positions 398 and 836 of the previously published sequence of hsp 65. The presence of amplified product was confirmed by agarose gel electrophoresis [11-13].
Restriction digestion and analysis
All steps of the test were carried out in accordance with the recommendations of the manufacturer (Salubris Inc.,Ttirkey). For BstE II digestion, 20µl of PCR product was added directly to a mixture containing 0.5µl ( ~ 5U) of enzyme, 2.5µl of restriction buffer, and the mixture was incubated for 60min at 2h. Similarly, 20µl of product was incubated for 18h at 37°C in a solution containing Hae III enzyme, the corresponding buffer .
Evaluation of restriction patterns
After digestion, 4µl gel loading buffer (3% orange G, gliserol in water) was added ,and 10µl of the mixture was loaded onto a Nusieve 3:1 agarose gel (FMC Bioproducts) and 8% polyacriylamdie gel. Fragments were visualized by staining with ethidium bromide. Products were evaluated with a NuSieve 3:1 agarose gel and polyacrilamid gel electrophoresis (8%). In the stage of evaluation, Mayker B and Mayker H (Salubris Inc., Turkey) molecular weight standards specific to enzyme restriction regions Bst E II and Hae III, that contains certain band size were used. 21 standard strains of mycobacteria were also analysed with this method and M.tuberculosis H37Ra was used as a control in every run [13].
Results
In the beginning stage of our study ,all steps in house PCR- RFLP method were evaluated comparatively with standardized Mycobacteria typing kit . In standardization procedure using reference strains, same PCR products (441bp) were obtained with both methods (Figure 1) obtained amplification product was cut by two separate restriction enzymes in accordance with reference literature and protocols of manufacturer. Final product were evaluated with 8% polyacrilamid gel electrophoresis and 4% Nusieve Agarose (Figure 2). While Hae III enzyme yielded similar results with both methods , with Bst E II enzyme band patterns inconsistent with algorithm table and results of procedure same enzyme was evaluated in comparatively with new products and at different concentrations. In all studies ,similar results were obtained with the same enzyme. Therefore, study was continued with a different standardized enzyme.
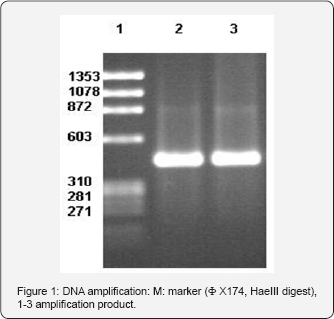
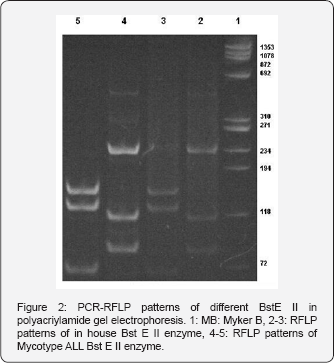
A total of 80 mycobacterial isolates were evaluated by using routine biochemical tests and PCR -RFLP. All the strains identified as M.tuberculosis by PCR-RFLP were identified as the same by biochemical tests and colony morphology. The 21 standard mycobacterial strains used as controls reproducibly produced RFLP patterns in accordance with the patterns previously reported in other studies. The number and rates of species isolated are shown in Table III. Of 80 mycobacteria strains, 70(87.5%) were identified as Mycobacterium tuberculosis complex and 4(5%) as Mycobacterium scrofulaceum, 3(3.75%) as Mycobacterium gordonae. 3(3.75%) strains displaying different restriction patterns could not be identified as they were found in the algorithm reported in the literature.
Discussion
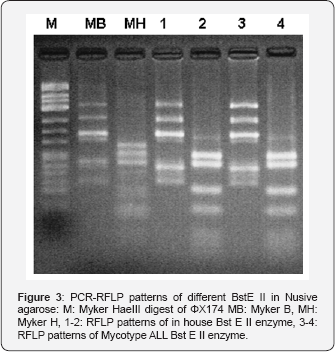
The resurgence of tuberculosis and apperance of multidrug- resistant strains of M.tuberculosis have intensified the need for the increased use of rapid methods for isolation and identification of mycobacteria from patients. Mycobacterial species identification methods depending on colony morphology, pigment formation and biochemical analysis are very cumbersome and require several week [2-20,21]. Commercially available probe based methods are limited to identification of only limited number of species and expensive. Methods based on DNA sequencing are not practical enough to apply routinely and require expensive equipment. PCR-RFLP is simple to perform, easy to read, and reproducible and is used in several laboratories to identify MOTT. Mycobacterial identification by PCR-RFLP depends on amplification and restriction enzyme analysis of hsp 65 gene ,which exist in all mycobacterial species but show sequence differences from one to the other [10-15] (Figure 3). These differences are identified by restriction enzymes that recognize and cut spesific DNA sequences. Obtained fragments are separated by electrophoresis and the size of fragments are compared to molecular weight marker. The most important stage in PCR-RFLP method is the enzyme cut of amplification product. In cases when enzymatic activity is low, this procedure cannot be performed in the expected period and misleading results may be obtained during evaluation. The activity of the enzymes used in the test varies with storage conditions employment period, heat during reaction and production characteristics. Therefore, in order to minimise any mistakes during evaluation, material to be used in all steps of the test should be controlled according to certain standards prior to the test. In the initial step of our study, MYCO TYPE ALL® (Salubris inc) kit was used as the standard for optimisation of the test and control of restriction enzymes [16]. Band patterns obtained with both tests were evaluated comparatively in algorithma [13,15,17]. While there was consistency between two methods in all steps of the test, in the evaluation made in in-house PCR -RFLP metho after cut with BstE II enzyme, different band patterns were identified in algorithma. Hence, study was continued with another enzyme the activity of which was controlled with the same method. After the cut procedure with enzyme, gel with suitable characteristics and concentration for the seperation of fragments should be used. Two methods used for this purpose are polyacrilamid gel (8-10%) and electrophoresis 4% NuSieve agarose. Both methods have certain advantages and disadvantages. The preparation of the former method is more practical and makes gel manipulation easier, but can not differentiate bands at similar magnitude. On the other hand, lather method enables detailed differentiation at suitable concentration and defines bands lower than 100bp clearly. It is disadvantage is that its preparation and gel manipulation is difficult.
Fragments in our study were evaluated with both methods. Although NuSieve agarose method was easier to employ, it was observed that it could not make differentiation as clear as polyacrylamide gel electrophoresis. In the evaluation stage of PCR- RFLP method, in order to differentiate species according to algorithm, the most important criteration is the selection of molecular weight markers containing suitable band size .The use of markers involving very far on very near band magnitude may cause incorrect evaluations. Therefore, the most commonly used molecular weight marker is ΦX174 Hae III DNA containing band sizes of 1353, 1078, 872, 603, 310, 281,271,234,194 ve 118 with this marker, especially after cut with BstE II enzyme , band sizes can be discriminated easily. Yet ,after cut with Hae III enzyme, misleading results may be obtained. Myker B and Myker H molecular weight markers just corresponding to the band sizes of at BstE II and Hae III enzyme cut regions are still at experimental stage. In our study, after amplification product were cut with BstE II and Hae III enzyme, we evaluated to band size by using three different marker (ΦX174 Hae III DNA, Myker B, Myker H). Band sizes especially smaller than 100bp and close to each other were readily established.
In group and type differentiation, reference algorithm and Mycobacteria typing kit were evaluated comparatively with the modified algorithm. While similar resutls were obtained in group differentiation by both algorithms, in type differentiation algortihm produced by the manufacturer yielded more detailed information. Of 80 mycobacteria isolate investigated in our study, 70(87.5%) was identified as Mycobacterium complex, 4(5%) as Mycobacterium scrofulaceum and 3(3.75%) as Mycobacterium gordonae. 3 isolates with different band patterns could not be defined by either algorithm.
Conclusion
In conclusion, PCR-RFLP method is a rapid, practical and reliable method that can be used routinely in laboratories for typing mycobacterium strains isolated from clinical samples. Restriction enzymes and molecular weight standards play an important part in the efficacy of the test. It is suggested that quality control of materials used should be made comparatively with standards before the test and that more than one molecular weight standards at proper band size should be used in order to minimise mistakes at the stage of evaluation.
References
- Primm TP, Ucero CA, Falkinham JO (2004) Health impacts of environmental mycobacteria. Clin Micribiol Rev 17(1): 98-106.
- Dunlap NE, Bass J, Fujiwara P (2000) Diagnostic standards and classificationof TB in adults and children. Am J Respir Crit Care Med 161(4): 1376-1395.
- Wayne LG, Sramek HA (1992) Agents of newly recognized or infrequently encountered mycobacterial disease. Clin Microbiol Rev 5(1): 1-25.
- Shojaei H, Heidarieh P, Hashemi A, Feizabadi MM, Daei Naser A (2011) Species identification of neglected nontuberculous mycobacteria in a developing country. Jpn J Infect Dis 64(4): 265-271.
- Banks J (1989) Treatment of pulmonary disease caused by opportunist mycobacteria. Thorax 44(6): 449-454.
- Crawford JT (1994) New technologies in the diagnosis of tuberculosis. Semin Respir Infect 9: 62-70.
- Hance AJ, Grandchamp B, Levy-Frebault V, Lecossier D, Rauzier J, et al. (1989) Detection and identification of mycobacteria by amplification of mycobacterial DNA. Mol Microbiol 3: 843-849.
- Springer B, Stockman L, Teschner K, Roberts GD, Bottger EC (1996) Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J Clin Microbiol 34(2): 296303.
- Vaneechoutte M, Beenhouwer HD, Claeys G, Verschraegen G, Rouk A D, et al. (1993) Identification of Mycobacterium species by using amplified ribosomal DNA restriction analysis. J Clin Microbiol 31(8): 2061-2065.
- Plikaytis BB, Plikaytis BD, Yakrus MA, Butler WR, Woodley CL, et al. (1992) Differentiation of slowly growing Mycobacterium species, including Mycobacterium tuberculosis, by gene amplification and restriction fragment length polymorphism analysis. J Clin Microbiol 30(7): 1815-1822.?
- Telenti A, Marchesi F, Balz M, Bally F, Bottger EC, et al. (1993) Rapid identification of Mycobacteria to species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol 31(2): 175178.
- Bahrmand AR, Bakayeva TG, Bakayev VV (1998) Use of restriction enzyme analysis of amplified DNA coding for the hsp65 gene and polymerase chain reaction with universal primer for rapid differentiation of mycobacterium species in the clinical laboratory. Scand J Infect Dis 30(5): 477-480.
- Kocagüz T, Yilmaz E, Ozkara Ç, Kocagoz S, Hayran M, et al. (1993) Detection of Mycobacterium tuberculosis in sputum samples by polymerase chain reaction using a simplified procedure. J Clin Microbiol 31(6): 1435-1438.
- Saifi M, Jabbarzadeh E, Bahrmand AR, Karimi A, Pourazar S, et al. (2013) Clin Microbiol Infect 19: 723-728.
- Devallois A, Goh KS, Rastrogi N (1997) Rapid identification of Mycobacteria to species level by PCR-RFLP analysis of the hsp 65 gene and proposition of an algorithm to differentiate 34 Mycobacterial species. J Clin Microbiol 35(11): 2969-2973.
- http://www.salubrisinc.com
- Ergin A, Kocagüz T, Us D (2000) Evaluation of 120 Mycobacterial strains isolated from clinical specimens to the species level by polymerase chain reaction-restriction enzyme analysis. Scand J Infect Dis 32(6): 657-662.
- Springer B, Stockman L, Teschner K (1996) Two laboratory collobrative study on identification of Mycobacteria : Molecular versus phenotypic methods. J Clin Microbiol 34(2): 293-303.
- Steringrube VA, Gibson JL, Brown BA (1995) PCR amplification and restriction endonuclease analysis of a 65- kilodalton heat shock protein gene sequence for taxonomic seperation of rapidliy growing Mycobacteria. J Clin Microbiol 33(1): 149-153.
- Silcox V (1992) Identification of mycobacteria. In: Isenberg HD, (Ed.), Clinical microbiology procedures handbook. American Society for Microbiology 3, Washington, DC, USA.
- Shenai S, Rodrigues C, Mehta A (2010) Time to identify and define non-tuberculous mycobacteria in a tuberculosis-endemic region. Int J Tuberc Lung Dis 14(8): 1001-1008.






























