A Study of Gastroenteritis Outbreak Caused by Aeromonas Verionii
Ostwal K1*, Dharne M2, Shah P3, Mehetre G2, Yashaswini D2 and Shaikh N3
1MIT Hospital, India
2National Collection of Industrial Microorganisms (NCIM), CSIR, India
3DrVM Government Medical College, India
Submission: February 01, 2017; Published: April 04, 2017
*Corresponding author: Karan Ostwal, Consultant Microbiologist MIT Hospital Aurangabad Maharashtra, India, Email: karanost55@gmail.com
How to cite this article: Ostwal K, Dharne M, Shah P, Mehetre G, Yashaswini D , et al. A Study of Gastroenteritis Outbreak Caused by Aeromonas Verionii. Adv 006 Biotech & Micro. 2017; 3(1): 555602. DOI:10.19080/AIBM.2017.03.555602
Abstract
Background: Aeromonas belongs to family Aeromonadaceae. Aeromonas verionii causes diarrhoea and gastroenteritis. Reports from Australia have suggested that there may be a connection between cases of Aeromonas-associated diarrhoea and the numbers of Aeromonas in the drinking-water. As, Aeromonas species is associated with water source, we hereby report an outbreak due to Aeromonas veronii in which source of infection was drinking water.
Aim: To determine the cause & identify the source of outbreak of gastroenteritis.
Material and methods: A cross- sectional study was performed in which seven patients with gastroenteritis were admitted to paediatric ward at Dr Vaishampayan memorial government medical college Solapur, Maharshtra during the month of october 2015. All the Patients gave history of fishing three days back to Sena river which lies at the border of Maharashtra & Karnataka. Patients suffered from severe acute gastroenteritis. Stool samples of all the seven patient were sent for microbiological examination. The organism was identified as Aeromonas verionii was subjected to antibiotic susceptibility testing by Kirby-Bauer disc diffusion technique. The source of infection was traced water sampling was done from the Sena river and A. verionii was isolated from water sampling. The isolates were sent for virulent gene detection & molecular typing.16S r DNA PCR and sequencing was done by ABI 3500xl genetic analyzer (Invitrogen).
Results: The Antibiotic Susceptibility pattern was identical of all the seven strains isolated from stool as well as the strain isolated from water sampling. The patients were treated with norfloxacin and were discharged after three days. However one of the strain isolated from stool sample and other one strain isolated from water sampling were confirmed as Aeromonas veronii biovar veronii by 16S rDNA PCR.
Conclusion: Early identification of the isolates and proper history can lead to rapid epidemiological investigations. This can aid in control an impending epidemic, if done vigilantly.
Keywords: Eno; Mesophilic; 16S r DNA PCR; Sequencing
Introduction
Aeromonas was classified previously with Vibrio species and Pleisomonas shigelloides in the Vibrionaceae [1]. Based on molecular genetic evidence, Aeromonas species is assigned separate family, Aeromonadaceae[1,2] . The genus aeromonas had been divided into 2 types- mesophilic which are potent human pathogens and psychrophilic consisting of non motile Aeromonas salmonicida [3]. Aeromonas verionii, is considered to be a food- borne bacterial pathogen causing infections animals and human being. It is generally found in fresh water.
A. veronii has been implicated in diseases like wound infections and diarrhoea. It is also found to be associated with septicemia in immune compromised patients. [4,5]. Aeromonas has been isolated from stools and reported with highest recovery during the warmer months [6]. The aeroslysin-hemolysin genes were found to cause diarrhoea in some patients who had A. verionii in their digestive system [7]. A numbers of factors, including age, immune competence, infection dose, underlying illness, and expression of sufficient virulence factors by the infecting organism, affect the ability of Aeromonas spp. to cause disease [8]. Reports from Australia have suggested that there may be a connection between cases of Aeromonas-associated diarrhoea and the numbers of Aeromonas in the drinking-water [9]. An outbreak is generally regarded when the case frequency is in excess of normal. The reporting of A. Verionii was never done in our region so we regard these cases as outbreak.
Materials and Methods
In cross- sectional study, seven patients with gastroenteritis were admitted to paediatric ward, with complaints of fever and diarrhoea since three days. The clinical picture of the seven patient is as mentioned in (Table 1-3).

Stool samples of all the seven patients admitted to paediatric ward were sent to department of microbiology. All stool samples were subjected to microscopic examination. Stool samples were inoculated on blood agar and MacConkey agar for culture and incubated at 37 °C for 16-18 hours. The culture strain was identified to genus level by using standard biochemical tests [3]. Antibiotic susceptibility testing was performed according to Kirby Bauer disk diffusion method by using antibiotic disk procured from Hi-media laboratories, Mumbai [10]. Two strains i.e one isolated from patient no.1 and the other isolated from water sampling were send to National Chemical Laboratory, Pune.
Water collection and sampling
The history given by all the patients was similar i.e, history of fishing in Sena river three days back, so a common source outbreak was suspected and source of infection was traced. Water sample was collected from the site where the children went for fishing. 100ml of water sample was collected from Sena river and was immediately cultured. Culture was done on double strength MacConkey broth was incubated for 48 hours [11]. After incubation subculture was done on blood and MacConkey agar. Molecular work-up- The genomic DNA isolation was isolated using Geneipure™ bacterial DNA purification kit (GeneipureTM) following manufactures protocol. The extracted genomic DNA was checked for integrity by 1% agarose gel electrophoresis in Tris-Acetate-EDTA buffer (40mM Tris, 20Mm acetate, 2mM EDTA) and stained using Gel red dye (BIOTIUM). [12] The concentration of the extracted DNA was checked using NanoDrop Lite spectrophotometer (NanoDrop Biotechnologies) [13]. Visualisation of DNA was done by gel documentation system (Proteinsimple) [14].
Amplification of 16S rRNA gene and enolase
Amplification of16SrRNAgenewas carriedoutusing universal bacterial primers 16F27 (5'CCAGAGTTTGATCMTGGCTCAG3') and 16R1525 (5'TTCTGCAGTCTAGAAGGAGGTGWTCCAGCC3') (Weisburg et. al., 1991) [15,16]. The PCR was carried out in 50μl reaction mixture containing 10nM (each) primer (Eurofins), 200μM (each) deoxynucleoside triphosphate (dNTP) (Genei), 1U of Taq polymerase (Genei) in the 10X reaction buffer, and 100ng of DNA extract as a template. The PCR conditions were 34 cycles of 30 s at 95 °C, 30s at 55 °C, and 1min 30s at 72 °C. Amplification of enolase gene was performed using enoF (5'ATGTCCAAGATCGTTAAAGTGAT3') and enoR 5'TTAAGCCTGGTTCTTCACTTCTT3' (Khajanchi et.al, 2010). [12] The PCR condition were 34 cycles of 30s at 94 °C, 30s at 55 °C, and 2min at 72 °C with final extention of 7min at 72 °C. PCR was performed in 50μl reaction mixture containing 10nM (each) primer (Eurofins), 200μM (each) deoxynucleoside triphosphate (dNTP) (Genei), 1 U of Taq polymerase (Genei) in the appropriate reaction buffer, and 100ng of DNA as a template. The amplified PCR products of 16S rRNA and enolase were purified using ExoSAP (New England BioLabs) and sequenced for in ABI 3500xl genetic analyzer (Invitrogen).
Phylogenetic analyses using 16S rRNA and enolase gene
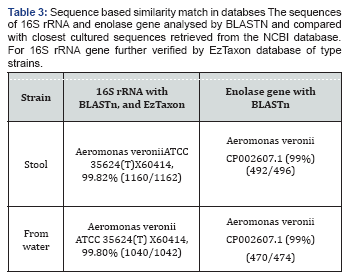
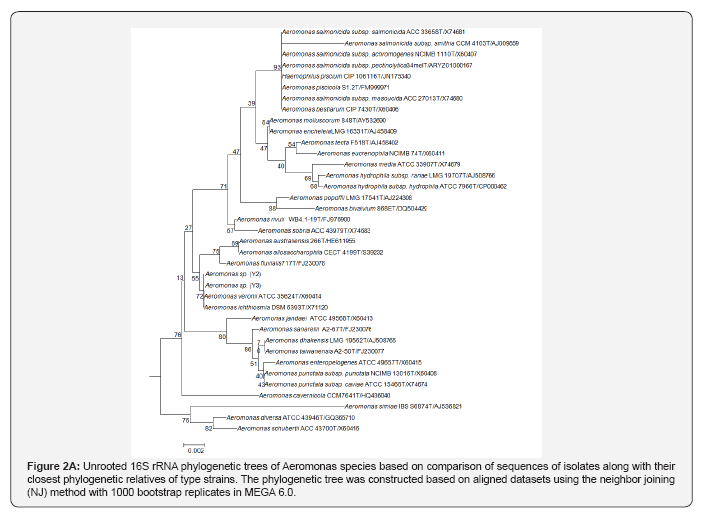
The obtained sequences of 16S rRNA and enolase of the isolates were edited using Chromas Lite (version 1.5) [17]. Edited sequencing data was analysed and cross-checked with BLASTN using default parameters [18] and compared with closest cultured sequences retrieved from the NCBI database for both 16S r RNA and enolase gene. For 16S rRNA gene, the species level identification was further verified by EzTaxon database of type strains with validly published prokaryotic names [19]. Phylogenetic tree for 16S rRNA was constructed by neighbor- joining method [20] using MEGA 6.0 program [21] to determine the relationship of these isolates with known sequences in database using 1000 bootstrap iterations. Based on enolase gene sequence phylogenetic tree was also constructed to determine the relationship with known sequences.
Result
Macroscopically, stool samples were semi-solid, greenish, non -adherent to bottle, no segments of worms were present.
Microscopy
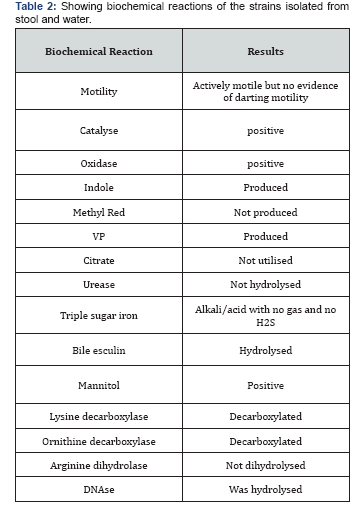
There was evidence of 4-5pus cells/hpf and actively motile bacteria in all the stool samples. There was no evidence of Red blood cells, ova, cysts and parasites. Culture characteristics- blood agar- beta-haemolytic colonies were observed. On MacConkey agar, non lactose fermenting colonies were found. Isolate from water sample- Culture characteristics- blood agar- confluent growth was observed MacConkey agar- nonlactose fermenting colonies were more in count than the lactose fermenting colonies. The non-lactose fermenting colonies were processed further. The biochemical reactions of all the seven strains isolated from stool and one isolated from water were identical and were as follows-
Antibiotic susceptibility test: All the isolates (seven from stool & one of water)-were sensitive to amikacin (23mm), norfloxacin (22mm), meropenem (24mm), cefotaxime (24mm), ceftazidime(19mm), piperacillin (25mm), amoxicillin + sulbuctam(21mm), cotrimoxazole (19mm) and resistant to ampicillin(8mm) and ciprofloxacin(10mm).
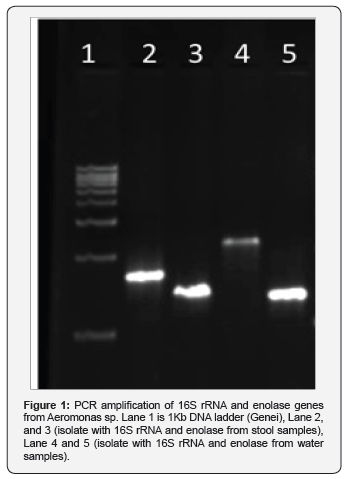
Molecular study: PCR amplification of 16S rRNA and enolase genes. The 16S rRNA amplicons (1.4 Kb) and enolase (1.3 Kb) genes were amplified to produce amplicons as shown in Figure 1.
Phylogenetic analysis based on 16S rRNA and enolase gene
The sequences were generated for two genes and for two isolates. The 16S rRNA gene sequence (1100bp) and enolase (1300bps) were used for phylogenetic analysis. Based on 16S rDNA similarity of the samples belonged to Aeromonas verionii ATCC 35624T as shown in Table 1. The enolase sequence showed 99% similarity to A.verionii ATCC 35624T from GenBank database (Figure 1). From the phylogenetic analysis of 16S rRNA and enolase gene, both the strains belong to A.veronii ATCC 35624T as shown in Figure 2A & 2B. The sequences generated in this study have been deposited in GenBank with accession numbers.

Discussion
A.verionii biovar verionii has rarely been isolated from humans and little information is available regarding its occurrence in clinical specimens and its ability to cause infections [22]. In India, Aeromonas associated diarrhoea has been reported from Bombay, Calcutta, Goa, Vellore, Pondicherry and Chennai,and the incidence in these areas was reported to range between less than 1% to about 13% [5]. Resistance is found mostly in aeromonas isolated from raw surface and ozonated water. In our study the isolate was found resistant to ciprofloxacin and ampicillin. Ampicillin resistance was also found in study by Taneja et al. The strain typing was done by biotyping and antibiogram in the same study. In our case apart from biotype and antibiogram, molecular work up was also done to establish the homology between the strains isolated from patients and environment. DNA homology was not associated in study done by, Moyer et al. [23] between gastroenteritis stool samples and drinking water samples. But in our case, DNA homology is present in water samples and stool samples. In some of the outbreak, the source of infection could not be traced [24].
Aeromonas is said to be pathogenic because it possess all the requirements of pathogenic bacteria. It attaches and enters into host cells through production of flagella, pili and adhesins including the presence of several virulence factors such as enolase [25]. Enolase catalyzes the reversible conversion of 2-phosphoglycerate into phosphoenolpyruvate. It is essential for the degradation of carbohydrates via glycolysis. The covalent binding to the substrate causes inactivation of the enzyme, and possibly serves as a signal for the export of the protein. Ottaviani et al. [26] reported the detected the alt gene in 76.4%and 55.5% of food and clinical isolates, respectively, and the ast gene in 16.4% and 61.1% of food and clinical isolates respectively in the isolates of children with diarrhea.
Hence we feel that early identification and molecular characterization of pathogenic strains of Aeromonas in patients as well as source are important steps in curtailing the spread and preventing further infections to occur. To the best of our knowledge, this is one of the rare article in which, tracing of source is done along with molecular processing.
Conclusion
Early identification of the isolates and proper history can lead to rapid epidemiological investigations. This can aid in control an impending epidemic, if done vigilantly. Limitation- The sequencing of the strains was performed in two cases only one from stool and other water sample.
References
- Winn C, Allen D, Janda M, Koneman W, Precop W, et al. (2006) Koneman's color atlas and textbook of diagnostic microbiology. (6th edn), Chap 7, Lippincott 2006, New York, USA, pp. 393-428.
- Colwell R, MacDonell R, De Ley J (1986) Proposal to recognize the family Aeromonadaceae fam. nov. International Journal of Systematic Bacteriology 36: 473-477.
- Collier L, Balows A, Susman M (1998) Bacterial meningitis. In : Patrick R Murray (Ed.), Topley and Wilson's Microbiology and Microbial infections. (9th edn), Chapter 17, Vol 3, Georgina bentliff, Great Britain, UK, pp. 299-306.
- Austin B, Austin DA (1999) Bacterial fish pathogens: disease in farmed and wild fish. Praxis Publishing, Chichester, UK, ISBN: 978-3-31932673-3.
- Alavandi S, Ananthan S (2003) Biochemical characteristics, serogroups, and virulence factors of aeromonas species isolated from cases of diarrhoea and domestic water samples in chennai. Indian journal of medical microbiology 21(4): 233-238.
- Le Chevallier M (1982) Aeromonas sobria in chlorinated drinking- water supplies. Microbial Ecology 8(4): 325-333.
- Arreola A, Hernande M Rodriguez C, Zuniga G, Figueras J, Garduno A, et al. (2007) Virulence potential and genetic diversity of Aeromonas caviae, Aeromonas veronii, and Aeromonas hydrophilia clinical isolates from Mexico and Spain: a comparative study. Canadian Journal of Microbiology 53(7): 877-887.
- Nichols G (1996) Health significance of bacteria in distribution systems- review of Aeromonas. London, UK Water Industry Research Ltd., (Report DW-02/A), UK.
- 9. Burke V (1984) Isolation of Aeromonas hydrophila from a metropolitan water supply: seasonal correlation with clinical isolates. Appl Environ Microbiol 48(2): 361-366.
- (2014) Clinical and Laboratory standards Institiute. M100-S23. Performance Standards for Antimicrobial susceptibilityTesting Twenty-Third Informational supplement; Pennsylvania, USA.
- Collee J, Fraser A, Marmio B, Simmons A (2007) Mackie and McCartney Practical Medical Microbiology. (14th edn), Elsevier Health Sciences, USA, pp. 95-113.
- Singh V, Chaudhary D, Mani I (2011) Molecular characterization and modeling of secondary structure of 16s-rna from Aeromonas verionii 3(1): 253-260.
- Li Y, Liu Y, Zhou Z, Huang H, Ren Y, Zhang Y, et al. (2011) Complete genome sequence of Aeromonas verionii strain B565. J Bacteriol 193(13): 3389-3390.
- Erova T, Fadl A Khajanchi B, Pillai L, Chopra A (2006) Mutations within the Catalytic Motif of DNA Adenine Methyltransferase (Dam) of Aeromonas hydrophila Cause the Virulence of the Dam-Overproducing Strain to Revert to That of the Wild-Type Phenotype. Infect Immun 74(10): 5763-5772.
- Shippy D, Fadl A (2014) tRNA Modification Enzymes GidA and MnmE: Potential Role in Virulence of Bacterial Pathogens. Int J Mol Sci 15(10): 18267-18280.
- Weisburg G, Barns M, Pelletier A, Lane J (1991)16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173: 697-703.
- Khajanchi BK, Fadl AA, Borchardt MA, Berg RL, Horneman AJ, et al. (2014) Distribution of virulence factors and molecular fingerprinting of Aeromonas species isolates from water and clinical samples: suggestive evidence of water-to-human transmission. Appl Environ Microbiol 76(7): 2313-2325.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI- BLAST: a new generation of protein database search program. Nucleic Acids Res 25(17): 3389-3402.
- Chun J, Lee H, Jung Y, Kim M, Kim S, et al. (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57(Pt 10): 2259-2261.
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4): 406-425.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12): 2725-2729.
- Mencacci A, Cenci E, Mazzolla R, Farinelli S, Francesco D, et al. (2003) Aeromonas veronii biovar veronii septicaemia and acute suppurative cholangitis in a patient with hepatitis B. JMM 52(8): 727-730.
- Moyer N (1992) Application of ribotyping for differentiating aeromonads isolated from clinical and environmental sources. Applied and Environmental microbiology 58(6): 1940-1944.
- Taneja N, Khurana S, Trehan A, Marwaha RK, Sharma M (2004) An outbreak of hospital acquired diarrhoea due to Aeromonas sobria. Indian pediatrics 41(9): 912-916
- Igbinosa IH, Igumbor EU, Aghdasi F, Tom M, Okoh AI (2012) Emerging Aeromonas Species Infections and Their Significance in Public Health. Scientific World Journal 2012: 625023.
- Ottaviani D, Parlani C, Citterio B, Masini L, Leoni F, et al. Putative virulence properties of Aeromonas strains isolated from food, environmental and clinical sources in Italy: A comparative study. Int J Food Microbiol 144(3): 538-545






























