16S rRNA Sequencing for Species Identification in Mixed Cultures for New Bio Preparations in Agriculture
Ostankova Yu V1, Semenov AV1, Pishchik VN2*, Popov AA3 and Totolian Areg A1
1Research Institute Pasteur, Russia
2Agrophysical Research Institute, Russia
3All-Russian Institute for Agricultural Microbiology, Russia
Submission: February 14, 2017; Published: March 24, 2017
*Corresponding author:Pishchik VN, PhD, Microbiology, Senior Researcher in Agrophysical Research Institute, Grazhdanskiy prospect 14, Saint Petersburg 195220, Russia, Tel: +7(812)534-45-65; Email: veronika-bio@rambler.ru
How to cite this article: Ostankova Yu V, Semenov AV, Pishchik VN, Popov AA, Totolian A A. 16S rRNA Sequencing for Species Identification in Mixed Cultures for New Bio Preparations in Agriculture. Adv Biotech & Micro. 2017; 2(5): 555599. DOI 10.19080/AIBM.2017.02.555599
Abstract
The study was carried out to identify bacterial strains in bacterial mixed cultures (or natural bacterial communities) with closely related bacteria isolated from plants rhizosphere and initially selected as a perspective cultures for biological preparation in agriculture. Phylogenetic relationships of these strains were determined by the comparison of the 16S rRNA sequences with the closest species from Gen Bank database. Bacteria were identified as Rhizobium galegae and Rhizobium leguminusarum (1st sample), Enterobacter sp. 638 and Enterobacter cloacae (2nd sample) Paenibacillus polymyxa, Paenibacillus peoriae and Microbacterium testaceum (3rd sample). Mycobacterium gilvum and Microbacterium testaceum (4th sample). It was concluded that 16S rRNA sequencing might be used to monitor the qualitative composition of bacterial associations.
Keywords: Mixed bacterial culture; Bacterial identification; Genotyping; Sequencing; 16 s RNA gene
Introduction
Based on the research findings that both synergistic interactions among bacterial species and the composition of the bacterial community are important in determining the level of ecosystem functioning [1], we proposed to use bacterial communities with different plant growth-stimulating and biological diseases control mechanisms to develop new bio preparations for agriculture. In this work there was a problem of isolation of pure bacterial culture from the mixed cultures (or natural bacterial communities) with closely related bacteria. In addition to that, the separation of natural microbial associations may leads to lose its activity. In this regard, is necessary to monitor the qualitative composition of bacterial associations to develop, to store and to use of biological preparations. We used the method of 16 S-rRNA sequencing for species identification in mixed bacterial cultures with closely related bacteria.
material and methods
Bacterial mixed cultures. We used 4 samples of bacterial mixed cultures. The samples 1 and 3 were mixed cultures; the samples 2, 4 were natural microbial communities. All bacteria were isolated from plant’s rhizosphere and Galega and Pisum plants nodules. Bacterial ribosomal RNA sequences by direct sequencing. Extraction of bacterial DNA was carried out in triplicate in three independently grown cultures. Extraction of DNA was carried out by the chloroform-saline standard method based on the lysis of cells and denaturation of cellular proteins with a solution that contains guanidine thiocyanate, and followed by ethanol precipitation of nucleic acids [2].
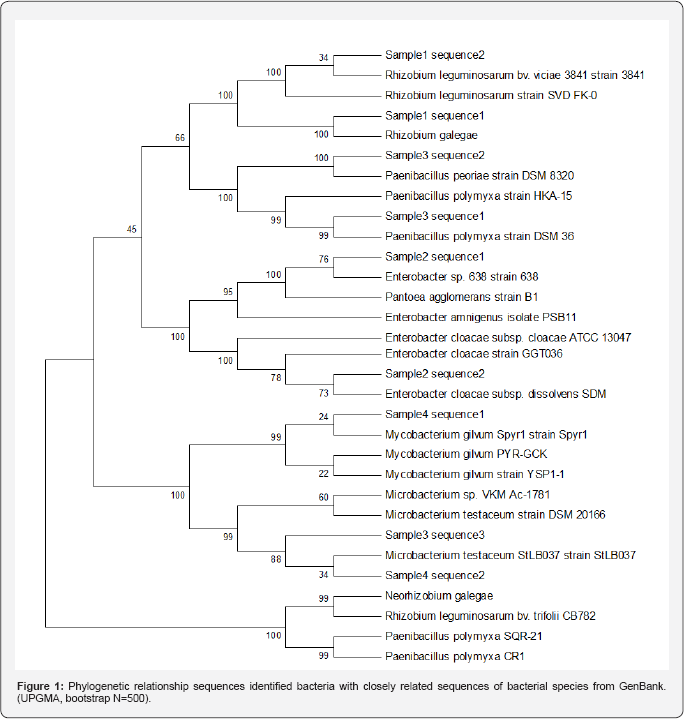
Random combinations and ratios of DNA concentrations of the following pure cultures: Leptospira interrogans, Stenotrophomonas maltophilia, Escherichia coli, Bordetella pertussis were mixed and used as a control. The "blind" method was used. For PCR were used two pairs universal primers flanking fragment about 1500bp. (F1 5’- AGAGTTTGATCMTGGCTCAG - 3’ and R1 5’- GGGGTATCTAATCCCGTTCG-3’, F2 5’- AACTTCGTGCCAGCAGC -3’ and R2 5’- GTCATCCCCACCTTCCTC -3’ or R2’ 5’- TACGGYTACCTTGTTACGACTT -3’). It was designed for the nucleotide sequenation of 16S rRNA (Figure 1).
PCR amplification was run in 25mcl amplification mixture with the primers 15pM each plus 67mM Tris HCl (pH 8.8), 16.6mM ammonium sulphate, 6.7mM MgCl2; 6.7mM EDTA; 10mM mercapto ethanol; 170 mg BSA; 1.0mM of each NTP; 1U of Taq DNA-polymerase (Fermentas). Denaturation (94 °C, 5min) was followed by 40 cycles of amplification: 94 °C-30sec, 55 °C-30sec, 72 °C-1min 20sec, final elongation 72 °C -7min. An additional method for the peparation of the amplificate products was used for accurate identification. A mixture of the amplificate products were separated on denaturing polyacrylamide gels according to standard procedures with modifications [3].
Electrophoresis was carried out in 8% polyacrylamide gel with a gradient of 45 to 80% (100% gel contains 7 M urea solution, and 40% deionized formamide solution). The gel was stained for 30min in Tris-acetate- EDTA buffer containing ethidium bromide, washed with deionized water, visualized under ultraviolet light. Fragments were excised from the gel
with amplification products and homogenized in 1.5ml tubes. 25ul buffer was added to the DNA elute and incubated for 15min at 37 °C. Then the mixture was frozen and thawed several times. Incubating was carried out overnight at 37 °C and centrifuged at 13k rpm for 10min. Purified fragments were used for sequencing reactions formulation using a set Genome Lab DTCS-Quick Start Kit (Beckman Coulter Inc., USA).
To analyze the reaction product was sequenced purified precipitate and dissolved in SLS-buffer containing formamide and placed in a genetic analyzer Genome Lab GeXP. The obtained sequences were compared with those available in the Gen Bank database. The primary analysis of the obtained sequences was performed using the NCBI. In each case, a comparison of sequences carried out on the three most identical sample nucleotide sequences with a match of at least 99%. Alignment of the nucleotide sequences was performed in the program MEGA version 5, using an algorithm CLUSTAL V [4].
Results and Discussion
The presence of two closely related bacteria Rhizobium galegae and Rhizobium leguminusarum was shown for the sample N1. The natural bacterial community (the sampleN2) consisted of closely related bacteria Enterobacter sp. 638 and Enterobacter cloacae. For the sample N3 the presence of three bacterial cultures, which sequences exhibited high 99% identity to the sequences of the following bacteria Paenibacillus polymyxa, Paenibacillus peoriae and Microbacterium testaceum wasshown. The bacteria Mycobacterium gilvum and Microbacterium testaceum were identified in natural bacterial community of the sample № 4. Take into accounts the data of 16S rRNA sequencing; we excluded Enterobacter cloacae strain (as a potential pathogenic bacterium) from the further work. The strain which exhibited high levels (99%) of DNA sequence identity with the Enterobacter sp. 638 needs further identification. It was concluded that 16S rRNA sequencing might be used to monitor the qualitative composition of bacterial associations to develop.to store and to use of Biological Preparations.
Acknowledgement
The authors gratefully acknowledge for the partial financial support to Mr. Zhemyakin SV (LTd Company "Petersburg Biotechnology".
References
- Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK (2005) The contribution of species richness and composition to bacterial services. Nature 436(7054): 1157-1160.
- Graham DE (1978) The isolation of high molecular weight DNA from whole organisms or large tissue masses. Anal Biochem 85(2): 609-613.
- Cremonesi L, Firpo S, Ferrari M, Righetti PG, Gelfi C (1997) Doublegradient DGGE for optimized detection of DNA point mutations. Biotechniques 22(2): 326-330.
- Higgins DG, Bleasby AJ, Fuchs R (1992) CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci 8(2): 189-191.






























