Evaluation of three DNA extraction methods of Mycobacterium tuberculosis DNA from Processed Sputum for testing by Three Real Systems
Gülnur Tarhan1* and ismail Ceyhan2
1 Department of Medical Microbiology, Adiyaman University, Turkey
2Ataturk Chest Diseases and Thoracic Surgery Education and Research Hospital, Turkey
Submission: November 27, 2016; Published: January 31, 2017
*Corresponding author: Gulnur Tarhan, Adiyaman University, Faculty of Medicine, Siteler Mahallesi, Ataturk Bulvar, No: 411 Adiyaman, Fax: +(90)4162231693; Tel: +(90)5059448568; Email: gulnur.tarhan@yahoo.com
How to cite this article: G#&252;lnur T, ýsmail C.Evaluation of three DNA extraction methods of Mycobacterium tuberculosis DNA from Processed Sputum for testing by Three Real Systems. 006 . Adv Biotech & Micro. 2017; 2(2): 555581. DOI:10.19080/AIBM.2017.02.555581
Abstract
Aim: The purpose of this study was to evaluate the efficiency of three DNA extraction method using three real-time PCR kits for direct detection of Mycobacterium tuberculosis in smear positive and negative sputum samples.
Background: Several Real Time PCR techniques are available for detection of Mycobacterium tuberculosis (MTB) in clinical samples.The success of final amplification and detection of nucleic acid depends on successful extraction of DNA from clinical samples.
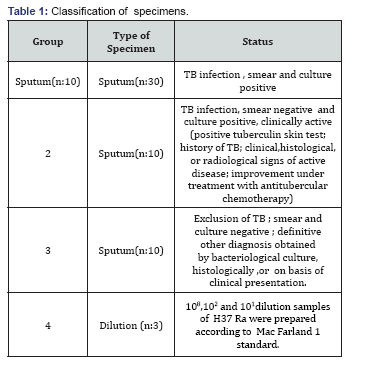
Materials and methods: 75 sputum samples (30 ARB+ , 30 ARB -, 15 healthy control, sputum samples ) and 3 dilution samples of H37Ra strain (108, 102, 101) were extracted using three extraction method (QIAamp Blood and Body Fluid Spin Protocol /Method A, Tris-EDTA (TE) boil extraction /Method B and Heliosis/ Method C). Real Time PCR tests were performed Icycler/ BioRad, TaqMa/ PE System 7700, Rotor Gene for each sample after all specimens were processed with NaOH- NALC decontaminaton method.
Results: The positive detection rate of I cycler system in smear positive sputum samples was 100 % for all extraction method. According to extraction method A,B and C; the positive detection rate of TaqMan 5700 and 7700 system in specimen Group 1 was 100%,87% ,90% and 100%,90% 100%, respectively. The positive detection rate of Icycler, TaqMan 5700 and 7700 for the extraction method A; B and C was 40 %,10% , 30% ; 30 %, 10% , 30% and 40 %,30% , 10%, respectively. For specimen Group 3, all positive detection resuts were negative of each extraction method by using three three real time PCR system. All PCR system results were positive for dilution 108 and 103 with each ectraction method. For 101dilution, Icycler and TaqMan 7700 system was negative for methot B
Conclusion: We concluded that all DNA extraction method can be used in RT-PCR to detect Mycobacterium tuberculosis from smear positive clinical samples. For smear negative sputum samples, Qiagen Body Fluids protocol and Heliosis method was found more sensitive than boiling method. There was no significant difference between the three -RT- PCR system in terms of diagnostic ability.
Keywords: DNA extraction; Real-time PCR; Tuberculosis y
Introduction
Pulmonary tuberculosis is a a highly contagious bacterial infection and major public health problem all over the world. The definitive diagnosis of Mycobacterium tuberculosis based on microbiology laboratory tests that are sensitive, specific, reproducible, and robust because of strong diagnosis evidence 1,2. Microscopic examination with either Ziehl-Neelsen (ZN)- or fluorochrome-stained smears is insensitive, detecting acid-fast bacilli only when there are 104 mycobacteria per ml.
Culture on solid medium is labor-intensive and too slow for clinical usefulness, taking a minimum of 2 weeks and often longer for microscopy-negative samples. Radiometric liquid (BACTEC) and biphasic (MB Chek) culture systems have improved both the recovery rates and the speed of isolation, but these systems still cannot influence initial bedside decision making 3-5. Nucleic acid amplification tests (NAATs) have a number of recognized advantages over other diagnostic assays, which include rapid and spesific diagnosis, compared to culture-based methods. A number of commercial NAAT systems and assays developed "inhouse" are currently in use for the detection of Mycobacterium tuberculosis and their use has seen many of the anticipated benefits of NAATs 6-8.
However, with increasing use of commercial and "in-house" NAAT systems over time and in different geographical settings, the need for both a considered approach to the application of NAATs and for an awareness of their limitations has arisen. These considerations include the sensitivity and specificity of the primary NAAT screening test, and where used, supplementary "confirmatory" tests as well as the prevalence of infection in various population groups 4,6,9,10. Within these methods, Real time PCR is commonly used for direct diagnosis of Mycobacterium tuberculosis from clinical specimens and to determine drug resistance. The presence of M. tuberculosis can be detected rapidly by measuring fluorescance signal simultaneously with amplification by means of this system 10,11. The sensitivity of the system varies according to DNA extraction from the sample and target specific primers and probes and commercial systems, employed for this are still at investigational stage. In these systems, whose control and amplification compounds have been standardized, there is no standardized method for DNA extraction. Therefore, various manual extraction methods and commercial kits are used 12.
The aim of this study was to evaluate three extraction methods of M. tuberculosis DNA from processed sputum: QIAamp Spin Column Kit (QIAGEN, Hilden, Germany), Tris-EDTA (TE) boil extraction 13 and Heliosis method (Metis, Inc,Turkey). The effectiveness of each extraction method was assessed using Icycler (BIO-RAD) , TaqMan 5700 and 7700 (Applied Biosystems) Real Time PCR systems.
Materials and Methods
Specimen groups
The performance of the three Real Time PCR (RT-PCR) system was evaluated using 30 ARB (+) and 10 ARB(-) patients diagnosed with active tuberculosis through clinical findings, radiology and laboratory investigations,and 10 healty controls and 3 dilution samples containing 108, 102 and 101 dilutions of H37Ra strain (overall 53 samples) were evaluated. Before the study, all clinical sample were verified with microscopy, culture and routine nucleic acid amplification test systems.
Specimen preparation
All sputum samples were decontaminated by treatment with an equal volume %3 NaOH trisodium citrate N-acetyl- L-cysteine (NALC) for 15 min at room temperature and were neutralized with sterile 0.067M phosphate buffer (pH: 6.8). After centrifugation at 3,000 Xg for 15 min, each pellet was resuspended in 1.0 ml sterile 0.067 M phosphate buffer (pH: 6.8) [14]. Three dilution sample was diluted 2.0 ml sterile 0.067 M phosphate buffer (pH: 6.8). 600 |il aliquot of the suspension was inoculated onto Lowenstein-Jensen (LJ) culture media and used for acid fast staining. Slopes was incubated at 37 °C. Slopes were inspected weekly for up to 8 weeks. Fixed smears were stained Erlich- Ziehl- Neelsen (EZN) staining [2]. All sputum samples were evaluated using Cobas Amplicor MTB and Gen Probe MTD during routine diagnosis. The remainder of aliquot was directly processed for RT-PCR or kept a-20 °C until used.
Dilution samples
A loopful bacteria were transferred to glass bead tubes containing 3 ml 0.9 % NaCl from 6 weeks culture of M.tuberculosis H37Ra strain. With 15 min vortex, bacteria was seperated as much as possible suspension prepared were adjusted according to Mac Farland 1 and 3X108 mycobacteria were yielded in 1 ml. Suspension was measured at 625 nm in spectrophotometre and adjusted so as to have a absorbance of 0.177. 1 ml of this suspension was mixed with 2 ml.0.9 % NaC and 1 ml suspension and 108 mycobacteria were obtained. From this main suspension, 108, 104 and 101 dilutions were obtained by serial dilutions. They were kept at -20 °C until used.
DNA extraction methods
QIAamp blood and body fluid spin column protocol : In this method ,samples were extracted according to Blood and Body Spin protocol of QIAamp, DNA Mini Kit. (QIAGEN,Hilden, Germany ). Briefly, 200 |il sample was transferred to clean microcentrifuge tube and 200 |il Buffer AL was added to sample. After it was mixed by pulse -vortexing for 15 sn, was incubated 56 ° C for 10 min. Briefly centrifuged the 1.5 ml in microcentrifuge tube to remove drops from the inside of the lid and 200 |il ethanol (96-100%) was added to the sample, and mixed again by pulse -vortexing for 15 s. After mixing , it was centrifuged in 1.5 ml microcentrifuge tube to remove drops from the inside of the lid.
The mixture was carefully applied to the QIAamp spin column (in a 2 ml collection tube) without wetting the rim, cap was closed ,and centrifuged at 6000 xg ( 8000 rpm) for 1 min. After centrifuging , the QIAamp spin column was placed in a clean 2 ml collection tube (provided) and the tube containing the filtrate was discarded. The QIAamp spin colum was carefully opened and 500 l Buffer was added AW1 without wetting the rim. The cap was closed and centrifuged at 6000xg (8000 rpm) for 1 min. Place the QIAamp spin column in a clean 2 ml collection tube (provided),and discard the collection tube containing the filtrate. The QIAamp spin column was carefully opened and added 500 l Buffer AW2 without wetting the rim. The cap was closed and centrifuged at full speed (20,000x g; 14 000 rpm) for 3 min. After centrifuged , the QIAamp spin column was placed in a new 2 ml collection tube and discarded the collection tube with the filtrate and it was centrifuged at full speed for 1 min. The QIAamp spin column was placed in a clean 1.5 ml microcentrifuge tube and discarded the collection tube containing the filtrate. The QIA amp spin column was openedcarefully and added 200 |il Buffer AE or distilled water. After It was incubated at room temperature for 1 min, centrifuged at 6000Xg (8000 xrpm)for 1min and kept at - 20 ° C until used [15].
Boiling methods
500 μl specimen was centrifuged and the pellet was washed twice with 500 μl of the TE buffer (10 mM Tris, 1 mM EDTA,? pH: 8.0) and then resuspended with 200 μl of same buffer .The samples were than incubated in a boiling water bath for 20 min ,centrifuged ,and the supernatants containing DNA were transferred to clean microcentrifuge tubes and kept at - 20 ° C until used. All centrifugation steps were performed at 14,000X g in plastic microcentrifuge tubes [13].
Heliosis methods
This method was performed according to the manufacturer's instructions supplied by the manufacturer ( Metis, Inc.,Turkey ). 500 μl DNA Lysis Binding solution was added to 100 μl sample. After vortexed, mixture was incubated 65 °C for 10 min and was kept at + 4 °C for 2 min. It was spin centrifuged, 500 μl DNA precipitation solution was added to it. Vortexed.It was centrifuged at 13.000Xg for 15 min.Supernatant was removed. 500 μl DNA Washing Solution was added and vortexed an centrifuged for 5 min at 13.000Xg. Supernatant was completely removed with pipette and dried at room temperature for 10 min. 20 ml DNA sample diluter solution was added, vortexed and spin centrifuged. DNA sample was kept at -20 ° C until study day[16].
Taq Man RT-PCR protocol
GATCTCGTCCAGCGCCGCTTCG-3') ve TCBP2 (5'- ACCGACGCCTA CGCTCGCAGG-3') primers, 163 bp
fragment of IS6IIO gene location on M.tuberculosis genome was amplified. Amplified product was detected by using specific fluorescent probes (FAM-GCTACCCAC AGCCGGTTA GGTGCTGGTG-TAMRA) For each sample, 5 μl of template DNA was incorporated into 45 μl PCR containing the amplification Tuberculosis Real Time PCR mix. All amplification stage was carried out ABI PRSIM 7700 SDS system .The optimized TaqMan RT-PCR protocol included initial step at 50 °C 2 min, 95 °C 10 min and was followed by a touchdown PCR protocol using the following conditions: 95 °C for 15 s, 61.5 °C for 1 min for 40 cylcles . The threshold cycle (Ct) value is the cycle at which there is a significant increase in fluorescence, and this value is associated with an exponential growth of PCR product during the log-linear phase. Test results were expressed according to Ct values computed comparativelywith positive and negative controls in amplificaiton graphics [17].
BIORAD RT-PCR protocol
In this system, Flurion- Mtbc QLS 1.0 M.tuberculosis detection kit was used.In amplificationprocedure, 163 bp fragment of IS6110 gene location on m.tuberculosis genome was amplified. Amplificaiton stage was carried out by BioRAD Icycler system. At this stage, after initial denaturation at 95 °C for 1min, overall 50 cycles were made 30 s at 95 μC, 30 s at 60 0C, 1 min at 72 μC , 30 s at 85 μC. Afterwards, it was kept waiting for 1 min 60 °C. PCR product amplificaition was visualised during reaction by fluoroescent SYBR- Green stain. In order to false positive result, melt curve stage was initiated after amplification. At this stage, overall 74 cycles were made 15 s at 60 - 950C and pirmer -dimer sepration was visualised during procedure [18] (Table 1).
Results
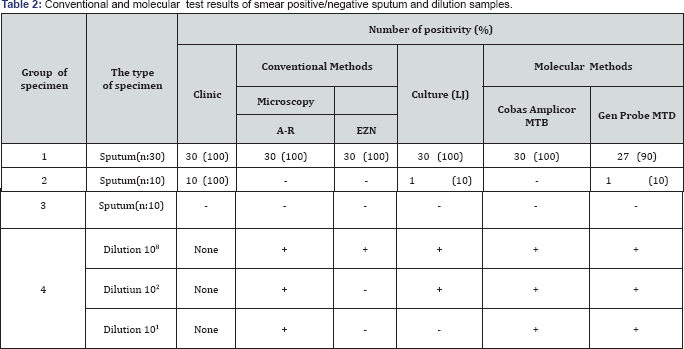
In the present study, clinical samples were evaluated with routine clinical,conventional methods ( microscopy, culture) and NAA methods (COBAS Amplicor MTB and Gen Probe MTD) before being evaluated with RT-PCR. The results of the study are illustrated in Table 2. All smear positive sputum samples were positive with microscopy, culture and Cobas Amplicor MTB molecular screening test. Three negative result was shown in smear positive sputum samples with Gen Probe MTD test. Microscopy positivity grade of these samples were +1 (Table 2). Of 10 smear negative sputum samples; microscopy (A-R and EZN) and Cobas Amplicor MTB test results were negative. Within the these samples, culture and Gen-Probe MTD test results were positive for one samples. Clinicical,radiological and histological findings of all smear negative samples were positive for TB (Table 2).
No positive or false positive result was observed in the control group (Group 3) or each sample, DNA extraction was performed by using Qiagen Body Fluids protocol (Method A), boiling (Method B) and Heliosis (Method C) extraction protocol. After DNA extraction, all samples were evaluated separately in Icycler (BIO-RAD Taqman (PE Biosystem) 5700 and 7700 systems. The positive detection rate of I cycler system. in specimen Group 1 was 100 % for all extraction method. According to extraction method A,B and C; the positive detection rate of TaqMan 5700 and 7700 system in specimen Group 1 was 100%,87% ,90% and 100%,90% 100%, respectively. The positive detection rate of Icycler, TaqMan 5700 and 7700 for the extraction method A; B and C was 40 %,10%, 30% ; 30 %, 10%, 30% and 40 %, 30%, 10%, respectively. For specimen Group 3, all positive detection resuts were negative of each extraction method by using three three real time PCR system. All PCR system results were positive for dilution 108 and 103 with each ectraction method. For 101dilution, Icycler and TaqMan 7700 system was negative for method B.
Discussion
Real time PCR is a commonly used molecular method to determined M.tuberculosis from clinical samples. This system is a commercially available system designed to decrease the time of the PCR by monitoring the amplification of the target sequences in real time by fluorescent probes [11]. The presence of M.tuberculosis can be detected rapidly by measuring fluorescance signal simultaneously with amplification by means of this system. The sensitivity of the system varies according to DNA extraction from the sample and target specific primers and probes and commercial systems, employed for this are still at investigational stage [7,11,12,19]. In these systems, whose control and amplification compounds have been standardised, there is no standardised method for DNA extraction. Therefore, various manual extraction methods and commercial kits are used. To increase the sensitivity of the system it is crucial to use the best method for extraction and purification af nucleic acids from clinical samples [16].
In this study, it was aimed to determine the most appropriate extraction methods in different RT-PCR studies from especially positive clinical samples. Each sample of DNA was extracted with Qiagen Body Fluids protocol, Boiling and Heliosis methods. After extraction procedure, all samples were evaluated separately in Taqman (PE Biosystem) 5700 / 7700 and Icycler (BIO-RAD) systems. When the sensitivity of the system was evaluated in all extraction methods, Mycobacterium DNA was detected at the rate of 90- 100 % in Icycler and TaqMan 7700 systems whereas this rate was 87- 100 % in TaqMan 5700 system for smear positive sputum samples. The lowest sensitivity was observed in the boiling method (Table 3).
Studies of clinical specimens conducted so far have differed in the type of PCR method used and also in the number and type of samples. A meta-analysis of papers on M. tuberculosis PCR has shown a sensitivity of <50% when PCR is performed on smear-negative pulmonary specimens, correlating with a smaller number of tubercle bacilli [7,12,20-23]. In this study, the sensitivity rates of Icycler, Taqman 5700 and 7700 system according to DNA extraction method A,B and C for smear negative sputum samples were; 10%,10%,30%; 30%,10%,30% and 40%,30%;10 % respectively. The positivity rate was very low for each system. The highest positivity was taken in Taqman 7700 system for Qiagen Body Fluids protocol (40%) (Table 3). Our results were similar to the others studies in smear negative samples. In all systems, false positive results were not recorded in smear negative sputum samples. Different results in all systems in the same extraction protocol did not suggest cross contamination. It was concluded that in order to obtain definitive data in the samples evaluation should be made with more samples.
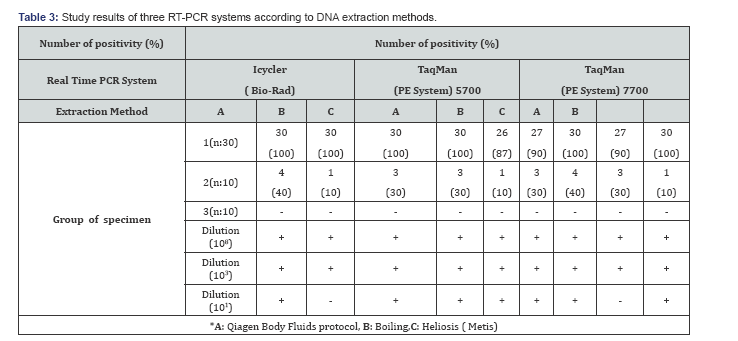
In the present study, three DNA extraction method was effective in smear positive sample. Qiagen Body Fluids protocol and Heliosis method was found more effective than boiling method. Three DNA extraction methods are fairly easy to perform, but the hands-on time is slightly different from one to another. Boiling methods has the shortest time of completion and the cheapest method. By making 10-fold serial dilutions of the DNA before amplification, the end points of detection was 101 dilution for DNA extraction method A and C in the three RT- PCR system.This result were negative for method B in the Icycler and TaqMan 7700 RT-PCR system.However, further testing using different sputum samples is necessary to confirm whether this observation is consistent regardless of variations in sputum matrices.
We concluded that all DNA extraction method can be used in RT-PCR to detect Mycobacterium tuberculosis from smear positive clinical samples. For smear negative sputum samples, Qiagen Body Fluids protocol and Heliosis method was found more sensitive than boiling method. There was no significant difference between the three -RT- PCR system in terms of diagnostic ability.
References
- Dunlap NE, Bass J, Fujiwara P, Hopewell P, Horsburgh CR, Salfinger M, Simone PM (2000) Diagnostic standards and classification of tuberculosis in adults and children. Am J Respir Crit Care Med 161: 1376-1395.
- Tuberculosis Division, International Union Against Tuberculosis and Lung Disease (2005) Tuberculosis bacteriology- priorities and indications in high prevalence countries: position of the technical staff of the Tuberculosis. Division of the International Union Against. Int J Tuberc Lung Dis 9(4): 355-361.
- Nolte FS, Metchock B (1995) Mycobacterium, In: Murray PR, Baron EJ, et al. (Eds.), Manual of clinical microbiology, (6th edn), American Society
- Watterson SA, Drobniewski FA (2000) Modern laboratory diagnosis of mycobacterial infections. J Clin Pathol 53(10): 727-732.
- Waard JH, Robledo J (2007) Conventional diagnostic methods. In: Palomino JC, Leao SC, et al. (Eds.), Tuberculosis 12: 401-424.
- Woods GL (2001) Molecular techniques in mycobacterial detection. Arch Pathol Lab Med 125(1): 122-126.
- Centers for Disease Control and Prevention (2000). Update: nucleic acid amplification tests for tuberculosis. Morb Mortal Wkly Rep 49(26): 593-594.
- World Health Organization (2010) WHO endorses new rapid tuberculosis test.
- Ling DI, Flores LL, Riley LW, Pai M (2008) Commercial nucleic- acid amplification tests for diagnosis of pulmonary tuberculosis in respiratory specimens: meta-analysis and meta-regression. PLoS One 3(2): e1536.
- Gibson UEM, Heid CA,Williams PM (1996)A novel method for real-time quantitative RT-PCR. Genome Res 6(10): 995-1001.
- Heid CA, Stevens J, Livak KJ, Williams PM (1996) Real-time quantitative PCR. Genome Res 6 (10): 986-989.
- MMWR (2009) Updated Guidelines for the Use of Nucleic Acid Amplification Tests in the Diagnosis of Tuberculosis 58(01): 7-10.
- Kocagöz T, Yilmaz E, Özkara Ç, Kocagöz S, Hayran M, et al. (1993) Detection of Mycobacterium tuberculosis in sputum samples by polymerase chain reaction using a simplified procedure. J Clin Microbiol 31(6): 1435-1438.
- Kubica GPW, Dye E, Cohn ML, Middlebrook G (1963) Sputum digestion and decontamination with N-acetyl-Lcysteine- sodiumhydroxide for culture of mycobacteria. Am Rev Respir Dis 87: 775-779.
- www.qiagen.com/literature/render.aspx
- Tarhan G, Ceyhan I, Çimçek H, Tuncer S (2009) Evaluation of the Efficacy of Five DNA Extraction Methods for the Detection of Mycobacterium tuberculosis DNA in Direct and Processed Sputum by an In-House PCR Method. Turk J Med Sci 39(2): 253-257.
- Applied Biosystems (2010) TaqMan® Universal PCR Master Mix, Instruction Manual.?
- BlORAD iCycler iQ™. Real-Time PCR Detection System. Instruction Manual. Catalog Number 170-8740.
- Kraus GT, Cleary N, Miller N, Seivright R, Young AK, et al. (2001)Rapid and specific detection of Mycobacterium tuberculosis using fluorogenic probes and real -time PCR. Mol Cell Probes 15(6): 375-383.
- Desjardin LE, Chen MD, Perkins l, Teixeria MD, Cave MD, et al. (1998) Comparison of the ABI 7700 system (Taqman) and competitive PCR for quantificaiton of IS 6110 DNA in sputum durung treatment of tuberculosis. J clin Microbiol 36(7): 1964-1968.
- Lachnik J, Ackermann B, Bohrssen A, Maass S, Diephaus C, et al. (2002) Rapid-cycle PCR and fluorimetry for detection of mycobacteria. J Clin Microbiol 40(9): 3364-3373.
- Armand S, Vanhuls P, Delcroix G, Courcol R, Lemaitre N (2011) Comparisonof the Xpert MTB/RIF test with an IS6110-TaqMan realtime PCR assay for direct detection of Mycobacterium tuberculosis in respiratory and nonrespiratory specimens. J Clin Microbiol 49(5): 1772-1776.
- Chang HE, Heo SR, Yoo KC, Song SH, Kim SH, et al. (2008) Detection of Mycobacterium tuberculosis complex using real-time polymerase chain reaction. Korean J Lab Med 28(2): 103-108.






























