Molecular Study of Vancomycin Resistance in Staphylococcus aures associated with Nosocomial Infections
Aalaa Aboelnour1*, Sherif M H El-kannishy2,3 and Maysaa E Zakil1
1Department of Clinical Pathology, Mansoura University, Egypt
2Department of Toxicology, Mansoura University, Egypt
3Department of Pharmacology and toxicology, University of Tabuk, Saudi Arabia
Submission: April 24, 2019;Published: September 11, 2019
*Corresponding author: Aalaa Abo elnour elmoursi, Department of Clinical Pathology, Mansoura University, Egypt
How to cite this article: Aalaa A, Sherif M H, Maysaa E Z. Molecular Study of Vancomycin Resistance in Staphylococcus aures associated with Nosocomial Infections. Trends Tech Sci Res. 2019; 3(3): 555614. DOI: 10.19080/TTSR.2019.03.555614
Abstract
Background:Staphylococcus aureus (S. aureus) causes hospital associated infections (HAIs).
Aim:The aim of the present study was to identify the emergence of vancomycin-resistant S. aureus among MRSA resistant andto identify the occurrence of van A, van B and van C genes among resistant isolates.
Method:The isolated strains confirmed to be S. aureus were subjected to full microbiological laboratory study for identification and antibiotics susceptibility beside molecular study for detection of vanA, vanB and van C genes by multiplex PCR.
Results:The study included 365 isolated S. aureus strains. Among isolated S.aureus strains, 113 (30.9%) was found to be MRSA. van A gene was recognized among 13 (68. 4%) resistant strains. van B was more commonly presents among resistant strains 17(89.5%).
Keywords: MRSA Multiplex PCR VanA VanB Vancomycin resistance
Introduction
Staphylococcus aureus is a leading pathogen in hospital acquired infections. It is isolated from various hospital acquired infections and its pathogenicity increased with the emergence of methicillin resistance (MRSA) in the last decades [1]. Vancomycin antibiotic is a glycopeptide antibiotic which have been considered a good therapeutic alternative for the treatment of MRSA. Unfortunately, resistant strains have been reported to reemerge among S. aureus species. The resistant strains have been reported to acquire thick wall preventing diffusion of vancomycin to the bacterial cells [2]. Vancomycin-resistant genes associated with S.aureus species are like those present in Enterococcus spp. These genes are seven types of resistance genes namely (vanA, B, C, D, E, G, and L). They are usually transferred from Enterococcus spp, by transposon Tn1546 [3].
The aim of the present study was to identify the emergence of vancomycin-resistant S. aureus among MRSA resistant strains and to identify the occurrence of vanA, vanB and van C genes among resistant isolates.
Materials and Methods
The study is a retrospective observational case series study that was conducted at Mansoura University Children hospital, Egypt from December 2014 till March 2016. The study included isolated S. aureus strains from children diagnosed to have health care associated infections (HCAI) according to CDC criteria of HCAI [4]. The patients signed written consents and the study was approved by Mansoura Faculty of Medicine ethical committee.
The isolated strains confirmed to be S. aureus by automated identification system Microscan (Bechman, USA), were subjected to full microbiological laboratory study including antibiotics susceptibility tests by disc diffusion method, manual determination of minimal inhibitory concentration for vancomycin and molecular study for detection of vanA, vanB and vanC genes by multiplex PCR.
Antibiotics susceptibility test
The used discs were vancomycin (30μg), erythromycin (15μg), ampicillin/sulbactam (20μg), amoxicillin/clavulanic (20/10μg), clindamycin (5μg)/, ceftriaxone 5μg, ceftazidime (30μg), cefoperazone (75μg), gentamycin (30μg), cefoxitin disc (30μg) (Oxoid Hampshire, England). Determination MRSA isolates was reported as those strains with inhibition zone ≤ 21 mm.
Broth Dilution Method of minimal inhibitory concentrations (MICs) for vancomycin
The determination of minimal inhibitory concentrations (MICs) for vancomycin was performed using standardized broth dilution techniques [5].
Vancomycin resistance among MRSA according to MIC was classified into susceptible, intermediate susceptible and resistant according to CLSI, 2009 [6].
Multiplex PCR for Van A, B, C genes Determination for MRSA strains
DNA preparation:One colony of pure culture was suspended in 25μL of sterile water and the suspension was put in the water bath at 100°C for 12 minutes. One micron of the suspension was used for PCR amplification.
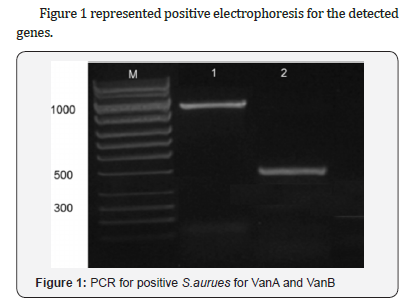
Multiplex PCR:The primers sequences used in PCR and amplification were as follow, vanA 5/-ATG AAT AGA ATA AAA GTT GC-/3, 5/-TCA CCC CTT TAA CGC TAA TA-/3 bp1032 [7], vanB 5/- GTG ACA AAC CGG AGG CGA GGA-/3, 5/-CCG CCA TCC TCC TGC AAA AAA-/3, 430bp [8], vanC 5/-ACG AGA AAG ACA ACA GGA AGA CC-/3, 5/-ACA TCG TGA TCG CTA AAA GGA GC-/3, 815bp [9].
The multiplex PCR was performed according to Perez-Roth et al. [10] using Qiagen amplification kit. Sterile distilled water was used as a negative control under complete sterile standard precautions for PCR.
After amplification 10μL of the reaction mixture was loaded onto a 1% agarose gel stained with 10μL ethidium bromide and electrophoresed to estimate the sizes of the amplification products with a 100-bp molecular size standard ladder (Sigma).
Results
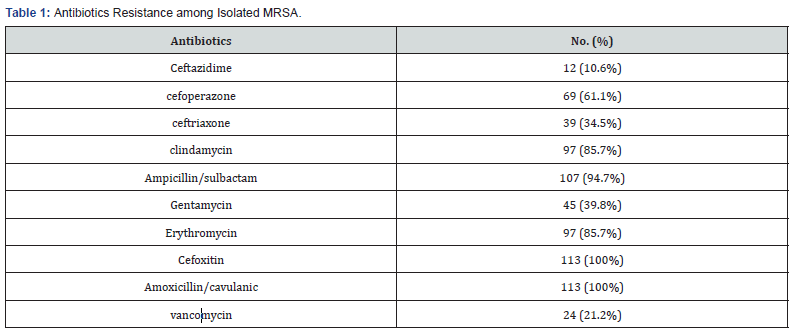
The study included 365 isolated S.aureus strains. Among isolated S.aureus strains, 113 (30.9%) was found to be MRSA, Isolated MRSA species were all resistant tocefoxitin and amoxicillin/ clavulanic with high resistance to ampicillin/sulbactam (94.7%), clindamycin and erythromycin (85.7%),vancomycin (21.2%). Table 1.
Vancomycin resistance among MRSA according to MIC was classified into susceptible, intermediate susceptible and resistant according to CLSI. VRSA was 19 (16.8%), VISA was 10 (8.8%) with MIC 4-8 μg/ml and susceptible strains were 84 (74.3%) with MIC 2μg/ml, data not shown.
Resistant vancomycin species was 15 strains with MIC 16- 32μg/ml, 3(2.7%) with MIC 64-128μg/ml, and one strain with MIC 256μg/ml, while VRSA was found in 10 isolates (8.8%).
Van A gene was detected among 3 (30%) isolates with intermediate susceptibility and in 13 (68.4%) resistant strains. vanB was more commonly associated with intermediate resistance pattern in 6 (60%) isolates and in 17(89.5%) resistant strains and none of the isolates had vanC, Table 2.

Discussion
The finding of the present study reported the presence of MRSA in 30.9% among HAIs in children hospital during the period of the study. The overall rates of MRSA in previous studies from Egypt were up to 70% [11,12]. While lower rates were reported in developed countries such as USA through implementing a multi model intervention including active surveillance, contact isolation, monitoring, and universal decolonization of patients in intensive care units [13]. The difference between our results and those from Egypt can be attributed to age of the included patients and the sample size.
Our findings demonstrated high resistance of MRSA to betalactams and macrolides antibiotics with rates from 85% up to 100%. These high rates of resistance are online by others reported from other studies [14, 15]. The high rate of resistance could be explained by the response of the MRSA strains to the selection pressure created by their constant exposure to antibiotics used in hospital settings [15].
In MRSA, 16.8% isolates were VRSA by determination of MIC with different MIC ranging from 16 to 512 Mg/ml.
In Middle East countries various studies have reported the presence of VRSA like Jordan [16], Saudi Arabia and Egypt [17].
In our study; about 20% of the isolates harbored at least one of the van genes. There is a possibility that these infections were caused by dissemination of a few clones of VRSA circulating in our hospital but, we can neither confirm nor exclude this possibility [18].
vanA gene was detected among 68.4% resistant strains and vanB was detected among 89.5% of VRSA strains. Similarly, vanA and vanB resistant genes were detected in 34% and 37% of clinical isolates, respectively [18]. The absence of van genes among VRSA strains are mainly due to the presence of other genes and mechanisms that attribute to the emergence of these strains in different proportions in VRSA
In this study, though we have found vanA and vanB genotypes among VISA isolates with high frequency 30% and 60% respectively. The presence of van genes A and B is considered among other mechanisms of VISA like thickened cell wall [2]. Patients infected with these strains usually have resistant pattern to vancomycin therapy when exposed to it. Moreover, the presence of carrier for these strains can be a source for emergence of VRSA isolates [19].
The findings highlight the emergence of vancomycin resistance among methicillin resistant S.aureus isolated from children with health care associated infections. Most resistant species revealed the presence of vanA and vanB as a responsible mechanism for this resistance.
Conclusion
The results of the current study illustrate the emergence of vancomycin resistance among methicillin-resistant S. aureus isolated from children with healthcare-associated infections. The majority revealed the occurrence of vanA and vanB as an accountable mechanism for this resistance.
References
- Valle DL Jr, Paclibare PA, Cabrera EC, Rivera WL (2016) Molecular and phenotypic characterization of methicillin-resistant Staphylococcus aureus isolates from a tertiary hospital in the Philippines. Tropical Medicine and Health 44: 3 DOI: 10.1186/s41182-016-0003.
- Liu C (2011) Clinical Practice Guidelines by the Infectious Diseases Society of America for the Treatment of Methicillin-Resistant Staphylococcus Aureus Infections in Adults and Children. Clinical Infectious Diseases 52: e18–e55. DOI:10.1093/cid/ciq146.
- Van Cleef BA, Monnet DL, Voss A, et al. (2011) Livestock-associated methicillin-resistant Staphylococcus aureus in humans, Europe. Emerg Infect Dis 17(3): 502.
- Jungk J, Como-Sabetti K, Stinchfield P, Ackerman P, Harriman K (2007) Epidemiology of methicillin-resistant Staphylococcus aureus at a pediatric healthcare system, 1991-2003. Pediatr Infect Dis J 26(4): 339-344.
- Tenover FC, Moellering RC Jr (2007) The rationale for revising the Clinical and Laboratory Standards Institute vancomycin minimal inhibitory concentration interpretive criteria for Staphylococcus aureus. Clin Infect Dis 44(9): 1208-1215.
- (2013) CLSI (Performance Standards for Antimicrobial Susceptibility Testing). CLSI Approved Standard M100-S23. Wayne, PA: Clinical and Laboratory Standards Institute.
- Donabedian S, Hershberger E, Thal LA, Chow JW, Clewell DB, et al. (2000) PCR fragment length polymorphism analysis of vancomycinresistant Enterococcus faecium. J Clin Microbiol 38(8): 2885-2888.
- Clark NC, Cooksey RC, Hill BC, Swenson JM, Tenover FC (1993) Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother 37(11): 2311-2317.
- Michel A, Richard Q (2001) Regulation of vanA and vanB type glycopeptides resistance in Enterococci. ACC 45(2): 375-387.
- Pérez-Roth E, Claverie-Martín F, Batista N, Moreno A, Méndez-Álvares S (2002) Mupirocin resistance in methicillin resistant Staphylococcus aureus clinical isolates in a Spanish hospital. Co-application of multiplex PCR assay and conventional microbiology methods Diag Microbiol Infect Dis 43(2): 123-128.
- El Kholy A, Baseem H, Hall GS, Procop GW, Longworth DL (2003) Antimicrobial resistance in Cairo, Egypt 1999-2000: a survey of five hospitals. J of Ant Chem 51(3): 625-630.
- Ahmed SH, Daef EA, Badary MS, Mahmoud MA, Abd Elsayed AA (2009) Nosocomial blood stream infection in intensive care units at Assiut University Hospitals (Upper Egypt) with special reference to extended spectrum βlactamase producing organisms. BMC Research Notes 2: 15- 19.
- Hadler JL, Petit S, Mandour M, Cartter ML (2012) Trends in invasive infection with methicillin resistant Staphylococcus aureus, Connecticut, USA, 2001–2010. Emerging Infectious Diseases 18(6): 917-924.
- Saderi H, Owlia P, Nadoushan MRJ (2009) Difference in epidemiology and antibiotic susceptibility of methicillin resistant and methicillin susceptible Staphylococcus aureus isolates. Iranian Journal of Clinical Infectious Diseases 4(4): 219-223.
- Kheder SI, Ali NA, Fathelrahman AI (2012) Prevalence and antimicrobial susceptibility pattern of methicillin resistance staphylococcus in a sudanese surgical ward. Pharmacology & Pharmacy 3(1): 103-108.
- Bataineh AB (2006) Resistance of Staphylococcus aureus to vancomycin in Zarqa, Jordan. Pak J Med Sci 22: 144-148.
- El-Banna TES, Sonbol FI, Aziz AA, El-Ekhnawy EAS (2015) Characterization of Vancomycin Resistant Staphylococcus aureusin Tanta University Hospital. Int J Curr Microbiol App Sci 4(10): 1-11.
- Saadat S, Solhjoo K, Norooz-Nejad MJ, Kazemi A (2014) VanA and VanB Positive Vancomycin-resistant Staphylococcus aureus Among Clinical Isolates in Shiraz, South of Iran. Oman Medical Journal 29(5): 335-339.
- Gardete S, Tomasz A (2014) Mechanisms of vancomycin resistance in Staphylococcus aureus. J Clin Invest Jul 124(7): 2836-2840.






























