Searching for Potential Entomopathogenic Bacteria in the Gut Microbiota of Halyomorpha halys
Samuel Cia1, Barbara Egger1 and Cosima Pelludat2*
1Fruit-Production Extension, Agroscope, Wädenswil, Switzerland
2Virology, Bacteriology and Phytoplasmology, Agroscope, Nyon, Switzerland
Submission: July 12, 2024; Published: July 22, 2024
*Corresponding author: Barbara Egger, Fruit-Production Extension, Agroscope, Wädenswil, Switzerland
How to cite this article: Samuel Cia, Barbara Egger and Cosima Pelludat*. Searching for Potential Entomopathogenic Bacteria in the Gut Microbiota of Halyomorpha halys. Agri Res& Tech: Open Access J. 2024; 28(5): 556423. DOI: 10.19080/ARTOAJ.2024.28.556423
Abstract
The brown marmorated stink bug, Halyomorpha halys, is an invasive, highly polyphagous pest of many agricultural crops, including fruit crops. To identify a potential biocontrol agent, the cultivable microbiota within the gut of 32 H. halys specimens collected during three sampling occasions from three different habitats in the Zurich region were examined. No culturable microbiota could be detected in 47% of the samples. Strains from the genera Pseudomonas and Enterococcus as well as the species Yokenella regensburgei and Acinetobacter johnsonii were the most abundant isolates. Due to the high beta diversity between the microbiota of H. halys specimens, no characteristic core microbiota could be determined. With the exception of one, all bacterial species that colonized the intestines of H. halys with a CFU higher than 104 are assigned to the biosafety level 2 and are therefore not suitable as biological control candidates.
Keywords: Halyomorpha halys; Swiss orchard; Screening; Gut; Culturable microbiota
Abbreviations: BCAs: Biological Control Agents; CFU: Colony Forming Units; NB: Nutrient Broth plates; MS: Mass Spectrometry; ARISA: Automated Ribosomal Intergenic 23 Spacer Analysis
Introduction
Halyomorpha halys Stål (Heteroptera: Pentatomidae), the brown marmorated stink bug, is native to subtropical and temperate regions in East Asia where it is widely distributed [1,2]. Today, this hemiptera is an invasive pest in North America, Europe and parts of South America [3]. First invasive H. halys populations were discovered in the US in 1996 [4]. Nowadays, H. halys is present in 26 European countries and considered to be widespread in regions of France, Italy, Portugal, Slovakia and Switzerland [5,3]. Halyomorpha halys is a polyphagous herbivore that feeds on more than 300 host plant species, including several commercial crops such as fruits, nuts, vegetables and ornamental plants [6]. Induced by its piercing-sucking feeding behavior, H. halys can provoke fruit deformation, discoloration and secondary fruit rot [4].
The control of H. halys is mainly based on the application of broad-spectrum insecticides with severe negative effects on non-target organisms and beneficials [7,8]. Moreover, H. halys is not sensitive towards most insecticides [8-10].
Alternative control strategies are therefore needed to protect crops from H. halys damage and to enable the sustainable cultivation of susceptible crops in infested areas. Utilizing biological control agents (BCAs) like entomopathogenic bacteria could present a viable alternative to the application of chemical insecticides. Some species belonging to the genera Bacillus, Burkholderia, Chromobacterium and Saccharopolyspora are commercially available BCA to control various insect pests [11]. Tozlu et al. [12] evaluated the efficacy of Bacillus, Brevibacillus, Pantoea, Pseudomonas and Vibrio strains against H. halys nymphs and found mortality rates of 75% to 100% under controlled conditions. The most efficient BCA were Bacillus cereus and Pantoea agglomerans. A Bacillus megaterium isolate from Palomena prasina (green shield bug) resulted in mortality rates up to 98% when tested against nymphs of the same species [13]. Screening the microbiota of H. halys to find bacteria with potentially harmful effects on the insect therefore seems to be useful to identify a suitable BCA. So far, studies on the microbiota of H. halys focused on the obligate, non-cultivable symbiont Candidatus Pantoea carbekii in the midgut gastric caeca (V4 region, [14]). Little is known about the microbiota present in the other gut regions (ventricles V1-V3) where digestion is mainly performed [15]. Aiming to deepen our understanding and to initiate the screening for potential entomopathogenic BCA, the culturable microbiota of the gut of H. halys was investigated in specimens originating from three different habitats in Switzerland.
Material and Methods
Collection of specimens
In 2020 a total of 32 H. halys adults were collected during three sampling events in the region of Zurich and Zug (Switzerland). Four specimens were captured within a hail protection net of an apple orchard in early May, where they were hibernating (=net samples). The other 28 individuals were collected from mid-May to early June, with 19 adults captured on ivy, Hedera helix (=ivy samples) and the 9 on sweet cherry, Prunus avium (=cherry samples) (Table 1). Collected H. halys were kept in ventilated plastic boxes at ambient conditions. If not dissected within 24 hours after sampling, parts of the last host plants were provided for nutrition. The dissection of the digestive tract of the adult H. halys was performed no more than seven days after their collection.
Dissection of the Digestive Tract
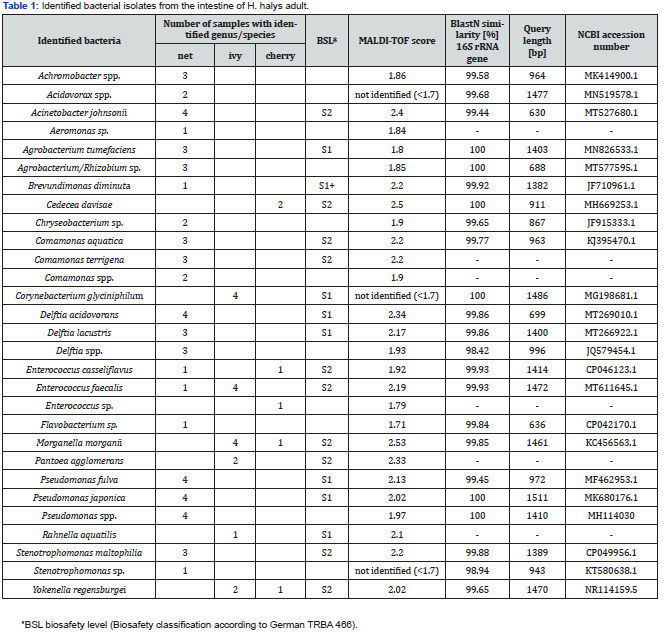
Prior to dissection, H. halys adult were kept at 4°C for at least 2 hours to immobilize the insects. Thereafter their head and legs were cut off with a sterile scalpel and the remaining body part was transferred on a sterile plate. The plate was placed in a UVCrosslinker (Vilber Lourmat UV-Crosslinker BLX-E254 6111 2081 1) for surface sterilization, and both insect’s ventral and dorsal body sides were each UV treated at 100V ~ 50 Hz for 30 seconds. The gut dissection was then performed under a dissection microscope. The abdomen was opened by cutting ventral alongside the tracheal openings with a sterile scalpel. Subsequently the dorsal part was lifted using sterile forceps. Immediately after opening, a few drops of 1x phosphate-buffered saline (PBS: 8 g/l NaCl, 0.2 g/l KCl, 1.44 g/l Na2HPO4, 0.24 g/l KH2PO4, pH 7,4) were pipetted onto the organs. The digestive tracts (including fore-, mid-, and hind guts but without the Malpighian tubes, Figure 1) were entirely removed and rinsed twice with PBS before being transferred into a 2 ml Eppendorf tube containing 500 μl PBS. The tubes were then kept at 4 °C until homogenization.
Halyomorpha halys Intestine Bacteria Isolation and Cultivation
The H. halys intestine was homogenized using a plastic pestle. After homogenization, the Eppendorf tubes were shaken at room temperature for 30 minutes at 1400 rpm followed by sonication (s 30 H, Elmasonic) for one minute at 45 kHz. For storage, additional 500 μl PBS were added to the homogenized intestine. Aliquots of 200 μl homogenized sample and 100 μl 40% glycerin were stored at - 80°C. A 10-fold serial dilution (up to 10-8) of each sample was performed, and 3 μl of each dilution was plated to determine the appropriate dilution for culturing approximately 20 to 200 colony forming units (CFU) on a nutrient plate when plating 100 μl per sample. The homogenized -80 °C samples were thawed, diluted accordingly and 100 μl of the final dilution plated on Nutrient Broth plates (NB (Difco), 8 g/l). In one case, plating 100 μl of the undiluted sample yielded less than ten colonies. This sample was consequently defined as “non-colonized”. Incubation occurred for at least 48 hours at 26 °C. Each colony of a plate was transferred with a sterile toothpick to a 96-well NB soft agar plate (7.5 g Agar/l). Morphologically identical colonies were grouped. The 96- well plates were incubated for 48 hours at 26 °C. After incubation, the bacterial isolates were transferred to a square NB medium plate of 120 x 120 mm using a 96-replicate plater and incubated for at least 24 hours at 26 °C before identification.
Identification of isolates
In a first attempt each bacterial isolate was analyzed using MALDI-TOF (Matrix Assisted Laser Desorption Ionization-Time of Flight; see paragraph below) mass spectrometry (MS). Isolates with a MALDI-TOF score ≥2 were identified at species level, those with a score between 2 and 1.7 at genus level [16]. Isolates whose MALDI-TOF score was below 1.7, were identified using 16S rRNA gene sequencing. The majority of MALDI-TOF identification results was confirmed by 16S rRNA gene sequencing using a representative isolate (Table 1). If more than three morphologically identical isolates were not identified by MALDI-TOF, three representative isolates were randomly selected for 16S rRNA gene sequencing. If the sequencing results led to identical identifications for the three samples, all isolates of this morphological group were assigned to the former identification. If the identification differed within the three representative samples, all unidentified isolates of the morphological group were sequenced. Colonies that failed to be identified by MALDI-TOF and 16S rRNA gene sequencing were defined as “not identified”.
MALDI-TOF MS
MALDI-TOF MS was performed as previously described [17]: cell material from a colony was smeared onto two spots of a MALDI target with a toothpick and each spot covered with 1 μl of matrix solution (1.5 mg of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile with 2.5% acetonitrile-water-trifluoroacetic acid, TFA). After air drying at room temperature the MALDI target was placed in the MALDI biotyper (Bruker Daltonics) and processed with the software FlexControl v3.0 (Bruker Daltonics).
PCR and Sequencing Reaction
For DNA extraction, a small loop of overnight cultured bacterial cell material was resuspended in 300 μl double-distilled water (ddH2O), heated at 98 °C for 20 min and centrifuged at 1400 rpm for 30 s. If not directly used as DNA template, the supernatant was removed and stored at -20 °C. For the amplification of the 16S rRNA gene coding region the primer pair 27F (5’-AGA GTT TGA TCC TGG CTC AG-3’) and 1492R (5’-CGG TTA CCT TGT TAC GAC TT-3’) (Weisburg et al. 1991) was used. A PCR reaction consisted of 10 μl Hotstar Taq Master Mix (Qiagen), 0.2 μl forward and 0.2 μl reverse primer (each 50 μM), 8.6 μl ddH2O and 1 μl template DNA. The PCR program steps were: 15 min 95 °C, 35 cycles of 30 s 95 °C, 30 s 56 °C, 90 s 72 °C and for the final extension 7 min 72 °C. The integrity and quality of the PCR products were verified by agarose gel electrophoresis (1% agarose gel, EtBr) and products purified using a MultiScreen PCR 96-well plate (Merck Millipore®, USA) following the manufacturers recommendation. Sequencing reactions with the BigDye® Terminator v1.1 cycle sequencing kit (Applied Biosystems) consisted of 2 μl BigDye® Terminator v1.1, 0.2 μl primer 27F or 1492R, 3.8 μl ddH2O, and 2 μl purified PCR product. Sequencing reactions were performed at 15 s 96 °C and 28 cycles of 15 s 96 °C, 15 s 45 °C, and 2 min 60 °C. Products were purified using the MultiScreen PCR 96-well plate cleanup kit. Sequencing and alignment were performed using a SeqStudio Genetic Analyzer (ThermoFischer) and the Geneious software (version 2020.0.4). For sequence analysis the basic logical alignment search tool (BLAST) [18] on NCBI (https://blast.ncbi. nlm.nih.gov/Blast.cgi) was used. Only if the similarity with the NCBI Blast sequence was higher than 99% and a unique match, an isolate was considered to be identified at species level, else it was defined at genus level.
Data Analysis
The bacterial CFU of an intestine determined by plating, was extrapolated according to the dilution level. The relative abundance was defined as the ratio between the CFU of a species to its genera in the intestine of a single individual and its total CFU (CFU/individual).
Results
CFU abundance in the midgut of H. halys samples
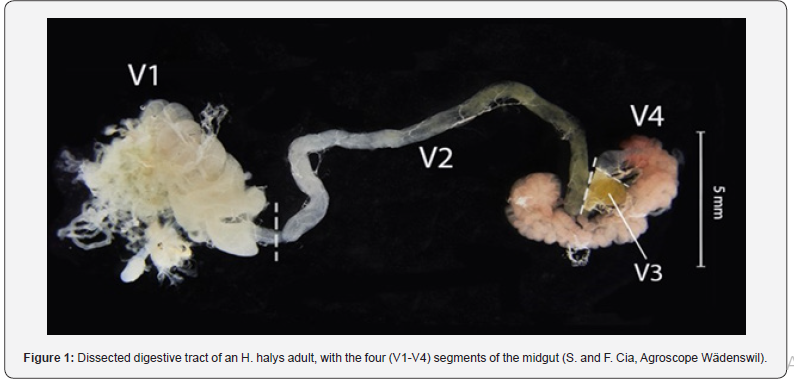
The analysis of the cultivated intestinal microbiota of 32 adult H. halys collected across three sampling events in the Zurich region revealed that no microorganisms could be cultivated under the given conditions in 15 (47 %) of these individuals (Figure 2). In the other 17 adults, the cultivable intestinal microbiota differed qualitatively and quantitatively between the three sampling events and among individuals. The absolute CFU of isolated bacteria revealed great variations between individuals, with a maximum of 5.2*108 CFU and a median of 5.9*104 CFU (Figure 2).
The Cultivated gut Microbiota of H. halys Samples
From the 17 adult H. halys with culturable microbiota a total of 816 bacterial isolates were cultivated. Of these isolates, 597 were identified to species or genus level by MALDI-TOF, whereas 148 were assigned by 16S rRNA gene sequencing. The remaining 71 colonies could not be identified. Identified bacterial isolates were assigned to 19 genera, 16 belonging to the phylum Proteobacteria, including the families of Enterobacteriaceae, Pseudomonadaceae and Moraxellaceae as the most prominent members. The remaining 3 genera belonged to the phyla Firmicutes, Actinobacteria and Bacteroidetes and were each represented by only one family, i.e. Enterococcaceae, Corynebacteriaceae and Flavobacteriaceae, respectively. Within the 19 identified genera, 18 species could be identified. For eight of the genera all cultivated colonies were assigned to a single species, namely Yokenella regensburgei, Corynebacterium glyciniphilum, Cedecea davisae, Morganella morganii, Pantoea agglomerans, Brevundimonas diminuta, Rahnella aquatilis and Acinetobacter johnsonii (Table 1).
Bacterial Diversity in the Cultivated gut Microbiota of H. halys Individuals
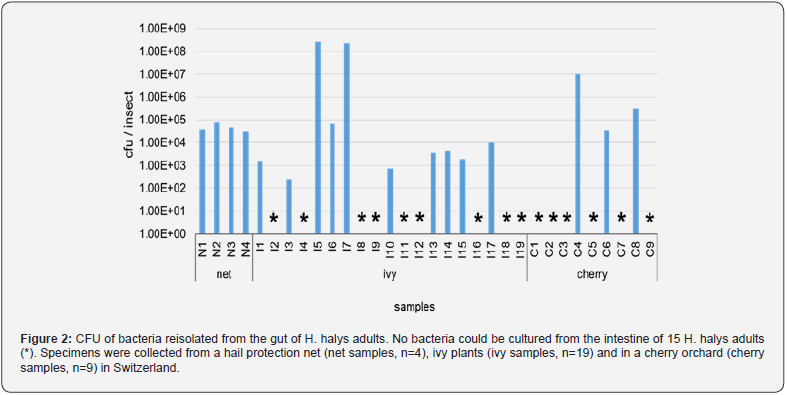
Overall, the samples were dominated by five bacterial genera. These genera could not be detected in all of the three sampled habitats. Acinetobacter spp., Chryseobacterium spp. and Pseudomonas spp. were identified in the net samples, whereas Y. regensburgei and C. davisae in the ivy and cherry samples. In all individuals, the relative abundance of the two most dominant genera accounted for at least 65 % of the cultivable bacteria, indicating a low alpha diversity.
In H. halys from the ivy and cherry samples, a single bacterial genus dominated the gut microbiota of an individual and accounted for at least 65 % of the relative abundance. In eight of these individuals, the relative abundance exceeded 99 % (Figure 3). In contrast to the ivy and cherry samples, the microbiota of H. halys individuals collected from hail nets was uniform and consisted of at least six different bacterial genera. Pseudomonas spp. and Acinetobacter spp. were the dominant genera in the net samples and accounted for 74 % of the determined CFUs. In total 13 genera were identified in the four individuals from the net sample, whereas only six and four genera were determined in the ivy and cherry samples, respectively, originating from the 19 individuals collected from ivy and the nine from cherry.
Only C. davisae, and Y. regensburgei reached more than 105 CFU within a single insect, the latter with a relative mean of 3.3 x 108 CFU. Y. regensburgei was isolated from three individuals, two of them collected from ivy the other one on cherry. Although M. morganii, P. agglomerans, P. japonica, C. glyciniphilum and A. johnsonii reached more than 104 CFU per insect, the majority of identified bacterial species remained below this level.
Except P. japonica, all bacterial species identified in the samples with >104 CFU/gut (e.g., Y. regensburgei, C. glyciniphilum, C. davisae, M. morganii, P. agglomerans and A. johnsoni) are classified as biosafety class 2 strains (S2, table 1). Cultivated bacteria of the biosafety class 1 comprised species such as Delftia acidovorans, D. lacustris or Rahnella aquatilis. Yet, they had all a relative abundance lower than 3*103 CFU/gut.
Overall, analysis of the culturable microbiota in the gut of adult H. halys from ivy and cherry revealed very low alpha but high beta diversity and predominantly S2-classified bacteria with a CFU >104. In contrast, the four net samples had a low beta - but higher alpha diversity than the ivy and cherry samples.
Discussion
The culturable gut microbiota of 32 adult H. halys collected from three different sampling events and habitats in central Switzerland was analyzed. The culturing approach provides information on the absolute and relative abundance of bacteria identified at genus and species level.
No microorganisms could be cultivated in 15 (47%) of the H. halys individuals. This result is consistent with a study conducted by Medina et al. [19], in which 54 to 73.5% of stink bug Nezara viridula adults had no detectable bacteria in the V1-V3 midgut ventricles, either by a culturing approach or by cultureindependent ARISA (Automated Ribosomal Intergenic 23 Spacer Analysis) analysis. Harada et al. [20] suggested that aphid guts generally lack gut bacteria and that bacteria detected in aphid guts are probably adventitious contaminants from the surface of host leaves or vascular tissues. The absence of a gut microbiota could be beneficial for H. halys in order to protect the extracellular symbiont in the V4 gut from harmful metabolites synthesized by ingested bacteria. The cultured microbiota of the samples analyzed may therefore represent a snapshot of bacteria that originate from recently ingested food sources and are largely eliminated once the digestive process is completed.
The cultured microbiota of the remaining 17 H. halys individuals varied in terms of bacterial genera and the total bacterial amount per individual. Proteobacteria were the most abundant, which is consistent with the finding that this phylum is dominant in almost all 28 insect families studied in a cross-taxon analysis that included Hemiptera [21]. In the presented study, Enterobacteriaceae were the dominant Proteobacteria, with Y. regensburgei and C. davisae as the most abundant species in the family. Alike, Yokenella sp. was reported as the most frequent bacteria species in a total of 173 N. viridula adults collected from soybean and eucalyptus trees in Argentina [19]. Enzymatic activities of Yokenella sp., Pantoea sp, and Enterococcus sp. in the V1-V3 region of N. viridula showed that these bacteria might have an impact on nutrition, detoxification and deactivation of chemical plant defenses, indicating that they may play a role in the fitness of their hosts [19].
The genera Pantoea and Yokenella could not be detected in the overwintering H. halys specimens collected from hail nets. On the other hand, dominant bacteria in the net samples, such as A. johnsonii and Pseudomonas sp., were not present in the ivy and cherry samples (Figure 3). In eight individuals, the intestinal microbiota were dominated by a single genus with a relative abundance of more than 99% (Figure 3). Low bacterial alpha diversity dominated by a single phylotype within insect individuals has also been reported in other studies [21,22]. This lack of diversity has been explained by the insects` ability to directly regulate the colonization and assembly of bacterial communities or by a negative interaction between the established symbiont and subsequent invading bacteria [21].
The analyzed microbiota show a high beta diversity among ivy and cherry H. halys individuals. In contrast, the four microbiota of H. halys from the net samples are quite uniform in terms of the identified bacterial genera and the bacterial abundances. This might be due to the long time the overwintering adults spent in the net, exposed to the same environmental conditions, deprived of food and/or with an impaired and less efficient immune system. The diversity of the bacterial microbiota found in the ivy and cherry samples could be explained by their different origin, as environmental conditions and food sources differed. However, the high mobility of active H. halys adults [23] prevents to establish direct correlations between the identified microbiota and food sources.
We assume that two essential prerequisites for an effective biological control agent against H. halys are the bacterium’s capacity to establish and multiply within the insect’s gut, and its classification as safe for both humans and the environment (S1 classification [24]). In line with this idea, entomopathogenic bacteria such as Serratia entomophila, the pathogen of the New Zealand warbler (Costelytra zealandica) or Paenibacillus larvae, the causative agent of a deadly honey bee brood massively colonizes the larval gut before initiating the killing step [25, 26]. In this study, all bacteria species, except P. japonica, having an abundance higher than 104 CFU/gut are classified at biosafety level S2 and are thus potential pathogenic or infectious organisms with a moderate hazard risk for healthy, adult humans [27].
Conclusion
The goal of this study was to identify culturable gut bacteria in H. halys that could potentially be used as BCA. The vast majority of the isolated bacteria are not suitable as biocontrol agents, because they are classified as potential pathogenic or infectious organisms for humans. However, further studies on antagonistic mechanisms against pentatomids and potential genetic similarities for host preference in opportunistic human pathogens and entomopathogens could be conducted with the isolated bacteria.
Acknowledgment
The authors would like to thank Lukas Sonderegger, Nicola Stäheli, Daniel Marzohl and Fabio Cia for their excellent support in the experiments and photography, Mitja Remus-Emsermann (Freie Universität Berlin), Diana La Forgia, Raphaël Groux and Patrik Kehrli (Agroscope Changins) for critically reading of the manuscript and helpful discussion, and Monika Maurhofer (ETH Zürich) for the supervision of the master thesis.
References
- Lee DH, Short BD, Joseph SV (2013) Review of the biology, ecology, and management of Halyomorpha halys (Hemiptera: Pentatomidae) in China, Japan, and the Republic of Korea. Env Entomol 42: 627-641.
- Zhu G-P, Ye Z, Du J, Dan Li Z, Ya-hui Z, et al. (2016) Range wide molecular data and niche modeling revealed the Pleistocene history of a global invader (Halyomorpha halys). Sci Rep 6: 23192.
- EPPO (2024) Halyomorpha halys (HALYHA). In: EPPO Glob. Database.
- Hoebeke ER, Carter ME (2003) Halyomorpha halys (Stal) (Heteroptera : Pentatomidae): A polyphagous plant pest from Asia newly detected in North America. Proc Entomol Soc Wash 105: 225-237.
- Haye T, Gariepy T, Hoelmer K, Jean PR, Jean CJ, et al. (2015) Range expansion of the invasive brown marmorated stinkbug, Halyomorpha halys: an increasing threat to field, fruit and vegetable crops worldwide. J Pest Sci 88: 665-673.
- Tracy C. Leskey, Anne L. Nielsen (2018) Impact of the Invasive Brown Marmorated Stink Bug in North America and Europe: History, Biology, Ecology, and Management. Annu Rev Entomol 63: 599-618.
- Leskey TC, Hamilton GC, Nielsen AL, Polk DF, Rodriguez S, et al. (2012) Pest Status of the Brown Marmorated Stink Bug, Halyomorpha Halys in the USA. Outlooks Pest Manag 23: 218-226.
- Leskey TC, Short BD, Butler BR, Wright SE (2012) Impact of the Invasive Brown Marmorated Stink Bug, Halyomorpha halys (Stål), in Mid-Atlantic Tree Fruit Orchards in the United States: Case Studies of Commercial Management. Psyche (Stuttg) 2012: 535062.
- Nielsen AL, Hamilton GC (2009) Seasonal Occurrence and Impact of Halyomorpha halys (Hemiptera: Pentatomidae) in Tree Fruit. J Econ Entomol 102(3): 1133-1140.
- Morehead JA, Kuhar TP (2017) Efficacy of organically approved insecticides against brown marmorated stink bug, Halyomorpha halys and other stink bugs. J Pest Sci 90: 1277-1285.
- Ruiu L (2015) Insect pathogenic bacteria in integrated pest management. Insects 6(2): 352-367.
- Tozlu E, Saruhan I, Tozlu G, Recep K, Faith D, et al. (2019) Potentials of some entomopathogens against the brown marmorated stink bug, Halyomorpha halys (Stål, 1855) (Hemiptera: Pentatomidae). Egypt J Biol Pest Control 29: 76.
- Aksoy HM, Tuncer C, Saruhan İ, Ismail E, Murat O, et al. (2018) Isolation and characterization of Bacillus megaterium isolates from dead pentatomids and their insecticidal activity to Palomena prasina nymphs. Akad Ziraat Derg 7: 21-28.
- Kenyon LJ, Meulia T, Sabree ZL (2015) Habitat Visualization and Genomic Analysis of Candidatus Pantoea carbekii, the Primary Symbiont of the Brown Marmorated Stink Bug. Genome Biol Evol 7(2): 620-635.
- Lomate PR, Bonning BC (2016) Distinct properties of proteases and nucleases in the gut, salivary gland and saliva of southern green stink bug, Nezara viridula. Sci Rep 6: 27587.
- Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G (2010) Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol 48(5): 1549-1554.
- Gekenidis M-T, Studer P, Wüthrich S, René B, David D (2014) Beyond the matrix-assisted laser desorption ionization (MALDI) biotyping workflow: in search of microorganism-specific tryptic peptides enabling discrimination of subspecies. Appl Environ Microbiol 80(14): 4234-4241.
- Altschul SF, Gish W, Miller W, et al. (1990) Basic local alignment search tool. J Mol Biol 215(3): 403-410.
- Medina V, Sardoy PM, Soria M, Carlos AV, Gabriel OG, et al. (2018) Characterized non-transient microbiota from stinkbug (Nezara viridula) midgut deactivates soybean chemical defenses. PLoS One 13: e0200161.
- Harada H, Oyaizu H, Ishikawa H (1996) A Consideration about The Origin of Aphid Intracellular Symbiont in Connection with Gut Bacterial Flora. J Gen Appl Microbiol 42: 17-26.
- Jones RT, Sanchez LG, Fierer N (2013) A Cross-Taxon Analysis of Insect-Associated Bacterial Diversity. PLoS One 8(4): 61218.
- Gauthier JP, Outreman Y, Mieuzet L, Simon J-C (2015) Bacterial Communities Associated with Host-Adapted Populations of Pea Aphids Revealed by Deep Sequencing of 16S Ribosomal DNA. Plos One 10(3): e0120664.
- Lee DH, Leskey TC (2015) Flight behavior of foraging and overwintering brown marmorated stink bug, Halyomorpha halys (Hemiptera: Pentatomidae). Bull Entomol Res 105(5): 566-573.
- Lelieveld HLM, Boon B, Bennett A, Brunius G, Cantley A, et al. (1996) Safe biotechnology. 7. Classification of microorganisms on the basis of hazard. Appl Microbiol Biotechnol 45(6): 723-729.
- Jackson TA, Boucias DG, Thaler J-O (2001) Pathobiology of Amber Disease, Caused by Serratia Spp., in the New Zealand Grass Grub, Costelytra zealandica. J Invertebr Pathol 78(4): 232-243.
- Yue D, Nordhoff M, Wieler LH, Genersch E (2008) Fluorescence in situ hybridization (FISH) analysis of the interactions between honeybee larvae and Paenibacillus larvae, the causative agent of American foulbrood of honeybees (Apis mellifera). Environ Microbiol 10(6): 1612-1620.
- Bundesamt für Umwelt BAFU, Bundesamt für Gesundheit BAG (2013) Einstufung von Organismen: Bakterien, Viren, Parasiten und Pilze.






























