RNA Sequencing Ixeris dentata var. albiflora
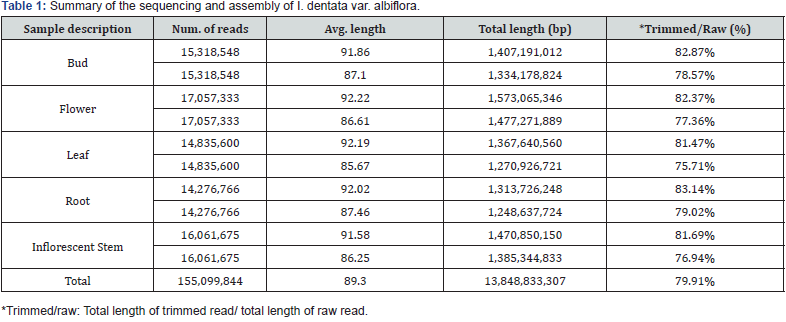
Sang-Hoon Lee1 and Yeon Bok Kim1,2*
1Department of Medicinal and Industrial Crops, Korea National University of Agriculture & Fisheries, Jeonju, Korea
2Department of Herbal Crop Resources, National Institute of Horticultural & Herbal Science, RDA, Eumseong-gun, Chungcheongbuk-do 369-873, Korea
Submission: January 19, 2024; Published: January 26, 2024
*Corresponding author: Yeon Bok Kim, Department of Medicinal and Industrial Crops, Korea National University of Agriculture & Fisheries, Department of Herbal Crop Resources, National Institute of Horticultural & Herbal Science, Korea
How to cite this article: Sang-Hoon Lee and Yeon Bok Kim*. RNA Sequencing Ixeris dentata var. albiflora. Agri Res& Tech: Open Access J. 2024; 28(1): 556400. DOI: 10.19080/ARTOAJ.2024.28.556400
dropdown Start here
Abstract
Ixeris dentata var. albiflora is a perennial herb, belonging to the Asteraceae family. I. dentata is distributed in East Asia specifically in Korea, Japan, China, and Mongolia. Their root and leaf are consuming as seasoned vegetables in Korea because of its bitter taste. As results of transcriptome using Illumina HiSeq 2000 sequencing platform, 107,178 transcripts with an average length of 1,281 nt and an N50 equaling 1,989 nt were identified from the 155,099,844 total clean reads after assembly. Assembled transcripts were further annotated by the BLAST similarity searches, Phytozome, NR, UniProtKB (Swiss-Prot), UniProtKB (TrEMBL) and TAIR ortholog identifiers. About 38-54% of transcripts were annotated. However, small numbers of genes were enriched with tissue specific manner: 141 in the floral bud, 150 in the flower, 137 in the root, 173 in the leaf and 161 genes in the inflorescent stem. Based on these transcriptome data, most of the tissue enriched genes were associated with phenylpropanoid and carotenoid biosynthesis in I. dentata var. albiflora. Our data may provide valuable information for the research on the functional genomics and pharmacological activities of I. dentata var. albiflora.
Keywords: Ixeris dentata; Next-generation sequencing; Illumina HiSeq; Transcriptome
Abbreviations: NGS: Next Generation Sequencing; BP: Base Pair; GO: Gene Ontology; CTAB: Cetyltrimethylammonium Bromide; FPKM: Fragments Per kb Per Million; FDR: False Discovery Rate; TAIR: The Arabidopsis Information Resource
Introduction
Ixeris dentata var. albiflora root and leaf are used as seasoned vegetables and has been used as traditional medicine to treat against mithridatism, calculous disease, indigestion, pneumonia, hepatitis, and so on. I. dentata var. albiflora is an undervalued crop, and mainly focused on the chemical component, medicinal effect and rarely studied the functional genomics. The initial step to provide data for functional genomics will generate transcriptome from the plant. NGS (Next Generation Sequencing) technologies such as Illumina’s Genome Analyzer, ABI’s SOLiD and Roche’s 454 GS FLX enable a new approach to genome sequencing, generating gigabases of sequence data in a few days for a few thousand US dollars [1]. For example, a single experiment on the instrument used in the present study (Illumina Genome Analyzer IIx, Illumina) can sequencing 225-250 million nucleic acid molecules, generating 45-50 Gigabases of 100 base pair (bp) paired-end sequence in roughly 9.5 days, where ‘paired-end’ refers to sequences obtained from the respective opposite ends of a single DNA molecule. ‘Short-read’ (36-72 nt) technologies such as Illumina and Applied Biosystems have proven to be exceptionally successful in a wide variety of whole-transcriptome investigations [2,3], but most of these studies have relied on prior sequence knowledge such as an annotated genome for qualitative and quantitative transcriptome analyses. Genome assembly of short sequences without any auxiliary knowledge has primarily utilized 454 sequencing data, due to the longer individual read lengths of 150-400 base pairs (bp). However, short-read sequencing (Illumina Genome Analyzer and solid) has been successfully used for de novo assembly of small bacterial genomes (2-5 Mbp), where 36 bp reads have been assembled and hybrid approaches used, whereas genomes are de novo assembled using a combination of reads from multiple sequencing platforms to overcome the inherent limitations of each technology, have been used to successfully assemble genomes up to 40Mbp [4,5]. In this study, the RNA-sequence information from I. dentata var. albiflora was obtained from Illumina paired-end sequencing technology. The sequencing data were subsequenctly assembled and annotated. Assembled unigenes were performed by the BLAST similarity searches and annotated with Gene Ontology (GO) identifiers. Our data may provide valuable information for the research on the functional genomics and pharmacological activities of I. dentata var. albiflora.
Materials and Methods
Plant materials
I dentata var. albiflora was grown at the experimental farm of the National Institute of Horticultural and Herbal Science, Rural Development Administration (Eumseong, Korea) in 2015. Different organs (flowers, inflorescent stems, leaves, bud and roots) were harvested from 15 plants in August 2015. Five plants for each replicate were used for biological repeats. All samples were immediately frozen in liquid nitrogen and then stored at -80°C for RNA isolation. The samples were ground into powder in a mortar with liquid nitrogen and total RNA was isolated separately using the modified cetyltrimethylammonium bromide (CTAB) method [6].
Library construction and Illumina sequencing
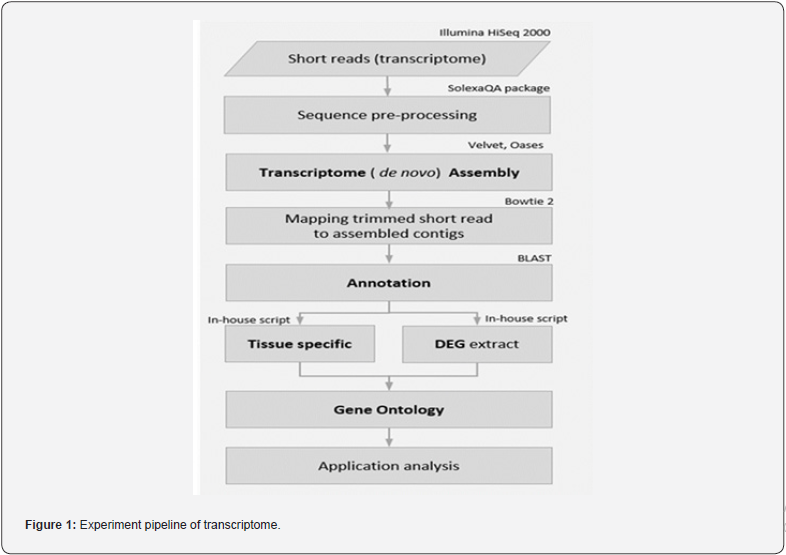
Total RNA samples were sent to Seeders inc. (Korea) for library construction, sequencing, data pre-processing and gene mapping. Magnetic beads with oligo (dT) were used to isolate mRNA from total RNA. Fragmentation buffer was then added to digest mRNA into short fragments that used as templates for cDNA synthesis using the TruSeq Sample Preparation Kit (Illumina, San Diego, CA, USA). Short fragments were purified using the QiaQuick PCR extraction kit and resolved with elution buffer for end reparation and single nucleotide A addition. Next, the short fragments related to adapters and analyzed using agarose gel electrophoresis to select suitable fragments for establishing a cDNA library by PCR. The resulting cDNA library was sequenced using the Illumina HiSeq™ 2000 system by solexaQA (Illumina, Inc., San Diego, CA, USA) package (Figure 1). Each sample was sequenced in duplicate.
De novo assembly and annotation
Clean reads were obtained, discarding adaptor sequences, unknown nucleotides larger than 5% (N ≥ 5%), and low quality reads. And then clean reads were assembled by using a program Velvet v1.2.08 [7] and Oases v0.2.08 [8]. We performed optimized k-mer search within the range of 35-85 for standard gene set assembly. Read mapping was performed by using bowtie2 v2.1.0 software [9]. Transcripts were submitted to protein databases, including the non-redundant (nr) protein database (http://blast.ncbi.nlm. nih.gov/), The Arabidopsis Information Resource (TAIR) database (https://www.arabidopsis.org/), Phytozome database (https:// phytozome.jgi.doe.gov/pz/portal.html), UniProtKB (TrEMBL and Swiss-Prot) database (http://www.uniprot.org/) for alignment and comparison using BLASTX with a significant E-value threshold ≤ 10-5. Blast2GO v. 2.5.0 (http://blast2go.com/b2ghome) was used for gene ontology (GO) annotation [10].
Analysis of DEGs and enriched gene selection
The fragments per kb per million reads (FPKM) method was used to calculate the level of unigene expression [11], since it can eliminate the influence of gene length and sequencing level on the calculation of gene expression [12]. DEseq2 software (Release 3.3) was used to detect DEGs [13]. Enriched genes were selected using the 2-fold change method and the false discovery rate (FDR) method also used simultaneously.
Results and Discussion
RNA sequencing and de novo assembly
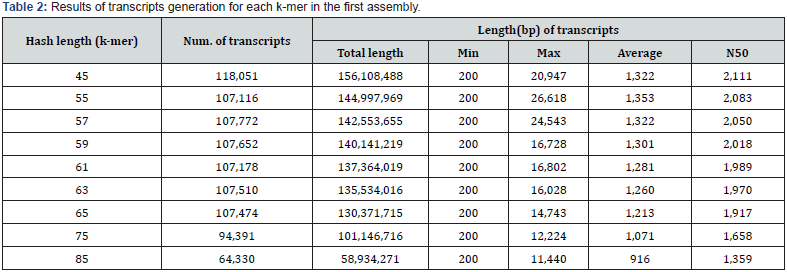
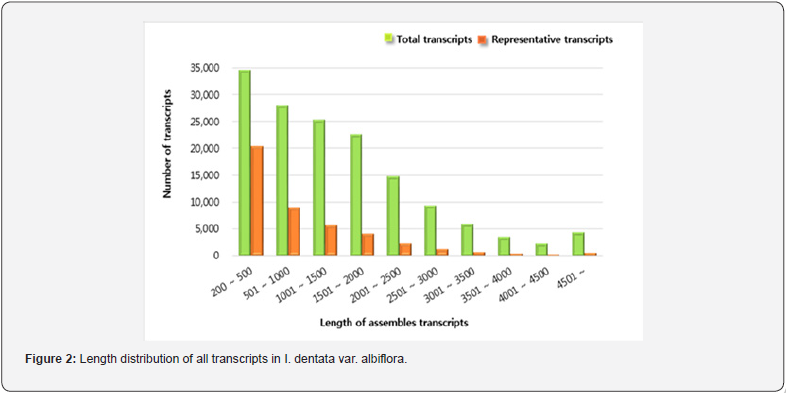
Five normalized libraries constructed from the RNA of I. dentata var. albiflora. After cleaning and quality measurement, a total of 13,848,833,307 clean reads (12.89 Gb) were generated with an average length of 89 nt (Table 1). Further, experiments for k-mer selection were performed. As a result, k-mer=59 and k-mer=61 was selected due to the small deviation from k-mer analysis (Table 2). The k-mer=59 and k-mer=61 showed very similar max length, Average length and N50 value. Final assembly results using k-mer=59 and k-mer=61, the length distribution of all transcripts was ranging from 200 nt to >4,500 nt (Figure 2). Specifically, 66.32% (29,177) of representative transcripts were found between 200 and 1000 nt, 21.86% (9,615) of representative transcripts in the range of 1000 to 2000 nt, 7.78% (3,424) of representative transcripts were between 2,000 and 3,000 nt, and 4.04% (1,776) were found more than 3,000 nt. Most transcripts generated from I. dentata var. albiflora were shorter than those of Chrysanthemum species [14]. Approximately, 50% of representative transcripts were found to be 200 to 1000 nt in I. dentata, whereas in Chrysanthemum species approximately 50% of the transcripts were found to be 600 to 1200 nt. This difference may be due to the sequencer platform because the transcripts produced from 454 GS FLX has –400 bp read length, while transcripts produced from HiSeq™ 2000 has -75 bp read length.
Transcripts annotation and analysis of DEGs
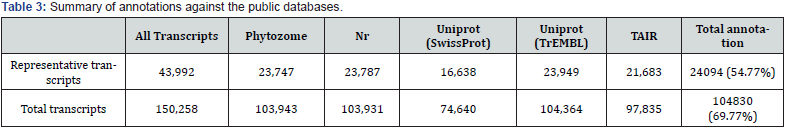
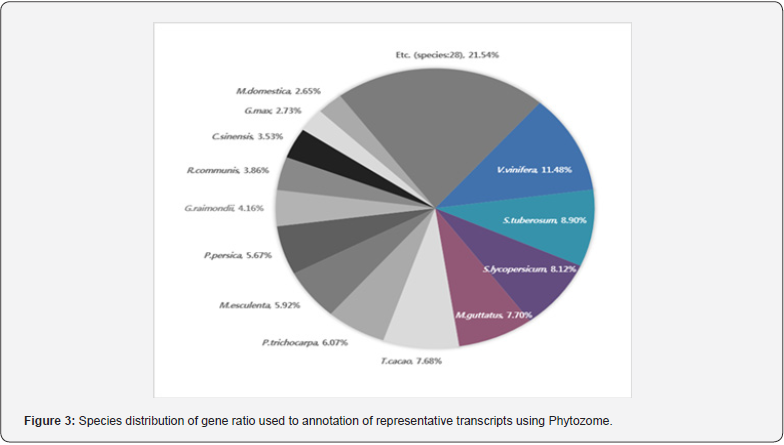
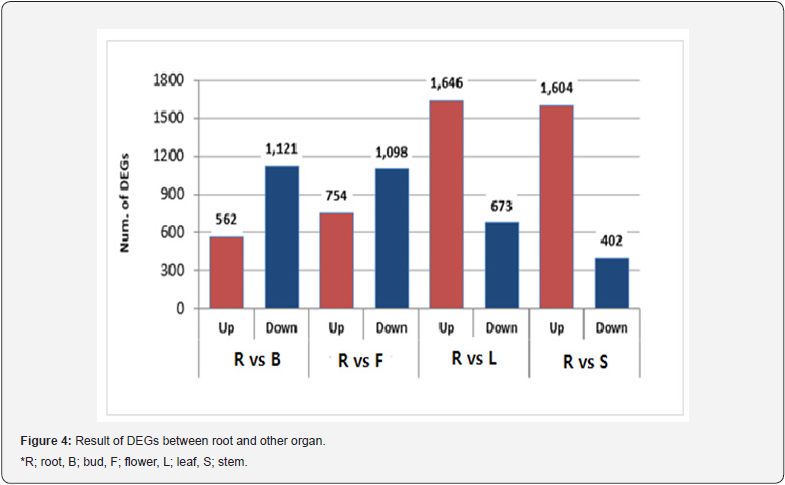
BLAST search against Phytozome, NR, UniProtKB (Swiss- Prot), UniProtKB (TrEMBL) and TAIR databases produced 23,747; 23,787; 16,638; 23,949; and 21,683 representative transcripts, respectively (Table 3). In Chrysanthemum species, the total annotated transcripts are 19,360 (46.4%) for C. boreale, 23,918 (41.9%) for C. morifolium and 43,550 (61.4%) for C. morifolium variety ‘Yuuka’ [15] The total annotated transcripts in this study were 24,094 (54.8%), reflecting similar percentage outcome. A total of 42 species representative transcripts were annotated using Phytozome program (Figure 3). Species distributions for the best match showed that the representative transcripts had the highest homology to genes of Vitis vinifera (11.48%), Solanum tuberosum (8.90%) and Solanum lycopersicum (8.12%). In a previous report using C. morifolium variety ‘Yuuka’ [15], the representative transcripts show the highest homology to genes from Vitis vinifera (26.8%), Solanum lycopersicum (16.8%) and Prunus persica (8.7%). Comparing I. dentata var. alblflora and C. morifolium variety ‘Yuuka’, they were highly homology to Vitis vinifera and Solanum lycopersicum. These results may be explained by the fact that both species are the same Asteraceae family. Using an FDR ≤ 0.001 and a log2 (fold change) ≥ 1 or ≤-1, this study identified significant DEGs between root and other part (Figure 4). Set root as control, the number of up-regulated genes in bud and flower were 562, 754 respectively, whereas the number of down-regulated genes were 1,121, 1,098 respectively. The number of up-regulated genes in the leaf and stem were 1,646, 1,604 respectively, whereas the number of down-regulated genes were 673, 402 respectively. The number of genes were up-regulated in the leaf and stem whereas in the flower and bud nearly few genes were up-regulated.
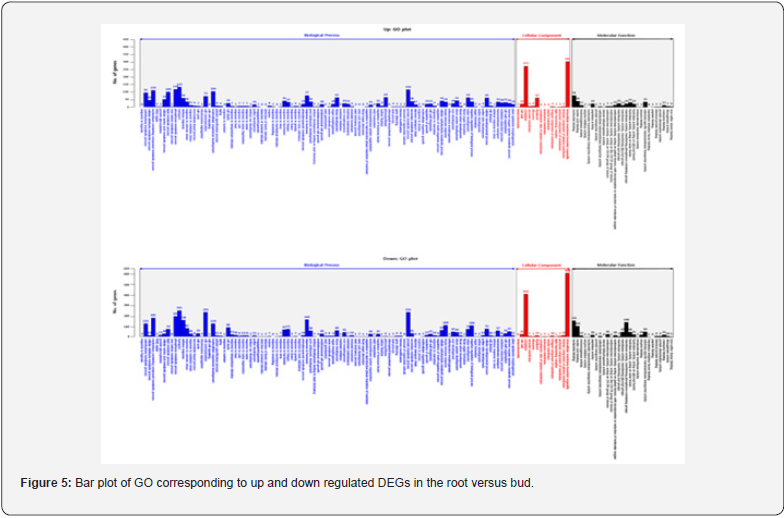
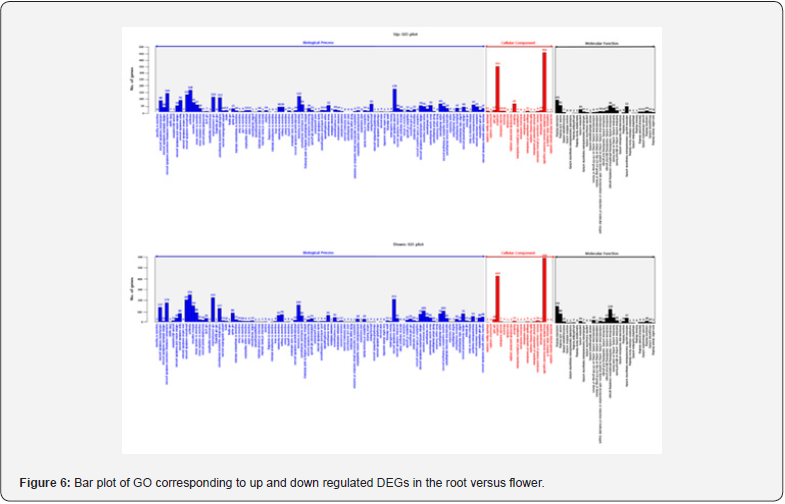
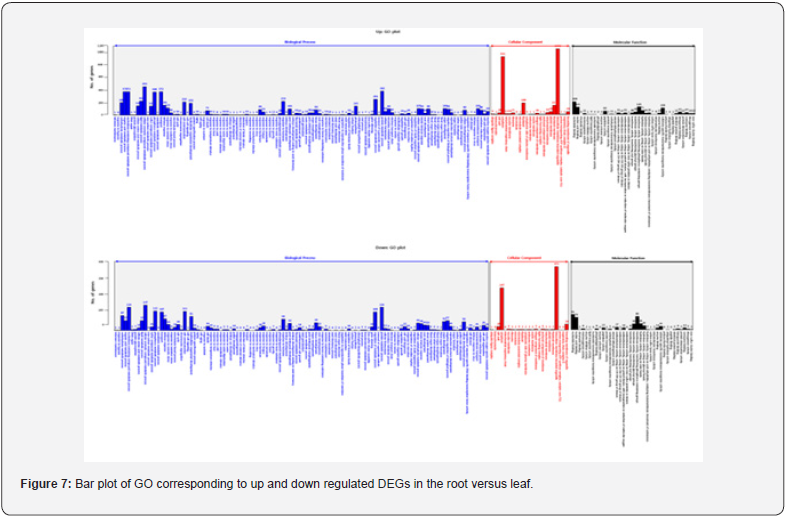
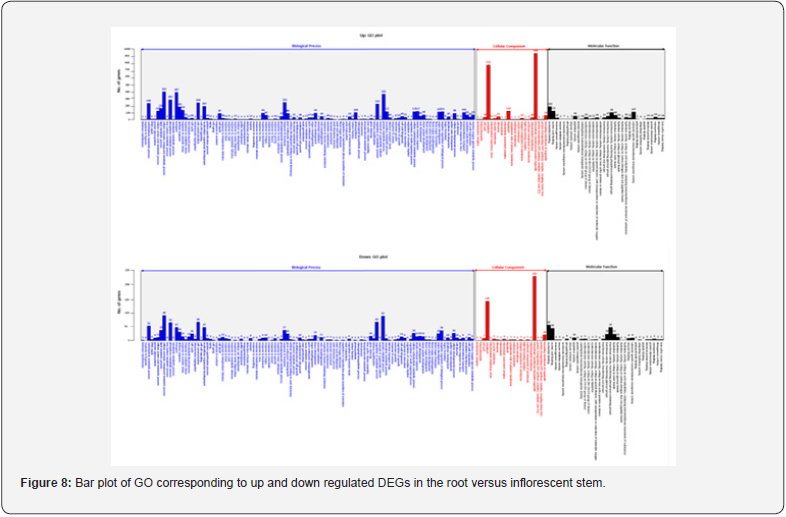
For GO analysis, DEGs were divided into three major categories: biological processes, cellular components, and molecular function. The results of the GO analysis of the up regulated DEGs and the down regulated DEGs through the DEGs analysis are as follows. In comparison of root and bud, the regulated gene was 2,044 in biological processes, 385 in cellular components and 381 in molecular function. Conversely, down regulated gene was 3,494 in biological processes, 502 in cellular components and 741 in molecular function (Figure 5). In comparison of root and flower, up regulated gene was 2,669 in biological processes, 966 in cellular components and 532 in molecular function. Conversely, down regulated gene was 3,678 in biological processes, 537 in cellular components and 746 in molecular function (Figure 6). In comparison of root and leaf, up regulated gene was 5,951 in biological processes, 2,592 in cellular components and 1,158 in molecular function. Conversely, down regulated genes were 2,327 in biological processes, 717 in cellular components and 501 in molecular function (Figure 7). In comparison of root and inflorescent stem, up regulated gene was 5,542 in biological processes, 2,084 in cellular components and 993 in molecular function. Conversely, the down regulated gene was 1,209 in biological processes, 417 in cellular components and 282 in molecular function (Figure 8). Compared with roots, flower and bud showed more down regulated genes, but leaf and inflorescent stem showed more up regulated genes. Taken together, GO classification of DEGs were found to be biological processes (62.5∼73.4%), cellular components (11.8∼25.0%), molecular function (12.1∼14.9%). In a previously reported Y. japonica study [Peng et al., 2014], GO classification was found to be biological processes (51.1%), cellular components (36.0%), molecular function (12.9%). Comparing I. dentata var. alblflora and Y. japonica, I. dentata var. alblflora, higher in GO term was cellular components.
Analysis of tissue enriched gene and select the target pathway gene.
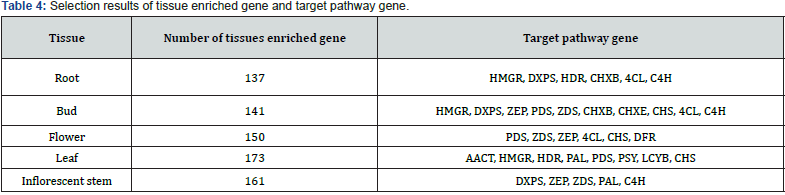
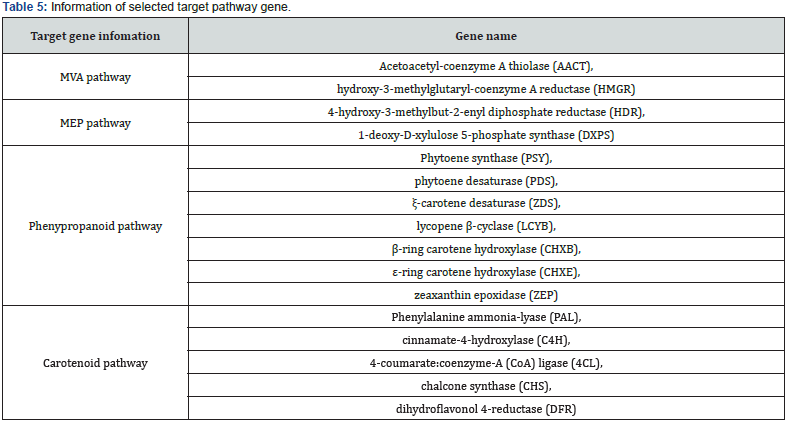
Using a log2 (fold change) ≥ 2 or ≤-2, this study identified significant tissue enriched gene between organs. As a result of tissue enriched gene analysis, the number of tissues enriched genes were found to be 173 genes in the leaf (22.7%), 161 genes in the inflorescent stem (21.1%), 150 genes in the flower (19.7%), 141 genes in the bud (18.5%) and 137 genes in the root (18.0%), respectively (Table 4). Further, targeted pathway genes were selected from each tissue. As a result, the number of targeted pathway genes were found to follow: 10 genes from the bud (HMGR, DXPS, ZEP, PDS, ZDS, CHXB, CHXE, CHS, 4CL and C4H), 8 genes in the leaf (AACT, HMGR, HDR, PAL, PDS, PSY, LCYB and CHS), 6 genes from the flower (PDS, ZDS, ZEP, 4CL, CHS and DFR), 6 genes from the root (HMGR, DXPS, HDR, CHXB, 4CL and C4H) and 5 genes from the inflorescent stem (DXPS, ZEP, ZDS, PAL and C4H), respectively (Table 4). Further examination was conducted to eliminate redundant genes and eventually selected pathway genes (Table 5). The total number of targeted pathway genes were found to be seven genes from the carotenoid pathway (PSY, PDS, ZDS, LCYB, CHXB, CHXE and ZEP) and five genes from the phenylpropanoid pathway (PAL, C4H, 4CL, CHS and DFR). Additionally, two genes from MVA pathway, (AACT and HMGR), and MEP pathway (HDR and DXPS) have been selected. The following experiment was carried out with two biosynthetic pathways (carotenoid and phenylpropanoid pathway) because of the abundance of selected target pathway genes. Based on these transcriptome data, most of the tissue enriched genes were associated with phenylpropanoid and carotenoid biosynthesis in I. dentata var. albiflora. Our data may provide valuable information for the research on the functional genomics and pharmacological activities of I. dentata var. albiflora.
Acknowledgement
This study was supported by the Cooperative Program for Agriculture Science & Technology Development (Project No. PJ01158102), Rural Development Administration, Korea.
References
- Holt RA, Jones SJ (2008) The new paradigm of flow cell sequencing. Genome Res 18(6): 839-846.
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ (2008) Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5(7): 613-619.
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Sci 320(5881): 1344-1349.
- DiGuistini S, Liao NY, Platt D, Robertson G, Seidel M (2009) De novo genome sequence assembly of a filamentous fungus using Sanger, 454 and Illumina sequence data. Genome Biol.
- Nowrousian M, Stajich JE, Chu ML, Engh I, Espagne E (2010) De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis. Plos Genetics 6(4): e1000891.
- White EJ, Venter M, Hiten NF, Burger JT (2008) Modified cetyltrimethylammonium bromide method improves robustness and versatility: the benchmark for plant RNA extraction. Biotech J 3(11): 1424-1428.
- Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18(5): 821-829.
- Schulz MH, Zerbino DR, Vingron M, Birney E (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28(8):1086-1092.
- Langmead B, Salizberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nature Methods 9: 357-359.
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M (2005) Blast2 GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18): 3674-3676.
- Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5: 621-628.
- Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7: 986-995.
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12): 550.
- Won SY, Kwon SJ, Lee TH, Jung JA, Kim JS, et al. (2017) Comparative transcriptome analysis reveals whole-genome duplications and gene selection patterns in cultivated and wild Chrysanthemum species. Plant Mol Biol 95(4): 451-461.
- Ren L, Liu T, Cheng Y, Sun J, Gao J, et al. (2016) Transcriptomic analysis of differentially expressed genes in the floral transition of the summer flowering chrysanthemum. BMC Genomics 17(1): 673.






























