The hta Gene of Thermus thermophilus HB8 Encodes a Protein with Homoserine Transacetylase and PHB Synthase Activity
Rigini M Papi* and Dimitrios A Kyriakidis
Laboratory of Biochemistry, Department of Chemistry, Aristotle University of Thessaloniki, Thessaloniki, Greece
Submission:August 23, 2025; Published: September 04, 2025
*Corresponding author:Rigini M Papi, Laboratory of Biochemistry, Department of Chemistry, Aristotle University of Thessaloniki, Thessaloniki, Greece
How to cite this article:Rigini M Papi and Dimitrios A Kyriakidis.The hta Gene of Thermus thermophilus HB8 Encodes a Protein with Homoserine Transacetylase and PHB Synthase Activity. Adv Biotech & Micro. 2025; 19(2): 556006.DOI:10.19080/AIBM.2025.19.556006
Abstract
A genomic survey of Thermus thermophilus HB8 for loci containing an α/β-hydrolase fold and the conserved lipase box motif identified the TTHA0759 locus. This gene was previously annotated as encoding a homoserine transacetylase (HTA), based on sequence homology to related proteins in other microorganisms. The hta gene was amplified from Thermus thermophilus genome and overexpressed in the mesophylic bacterium of Escherichia coli. The resulting recombinant protein, rHTA, exhibits dual functionality as both a homoserine transacetylase and a PHA synthase, as both activities involve the transfer of an acyl moiety. Kinetic studies showed that rHTA had a Km value of 0.11 mM for acetyl-CoA and a Km for synthase of 0.25 mM with 3HBCoA as substrate. Both Km values are very close to the native enzymes. Synthase activity is activated by acetyl-CoA, acetylphosphate and inhibited by CoA, NAD, NADP, similarly to the native enzyme. Treatment of rHTA with alkaline phosphatase leads to loss of its enzymic activity. In addition, synthase activity is inactivated by iodoacetamide suggesting that a cysteine moiety was involved in the catalytic reaction. Interestingly, the recombinant protein was found to catalyze the in vitro polymerization of (R)-3-hydroxybutyryl-CoA into polyhydroxybutyrate (PHB), demonstrating a high affinity for PHB granules. Notably, deletion of its transmembrane domain abolished this granule-binding capability, indicating that membrane association is critical for its interaction with PHB. These findings suggest that T. thermophilus HB8 utilize the homoserine transacetylase as a key component in the biosynthetic pathway of polyhydroxyalkanoates, facilitating their accumulation as intracellular storage biomolecules.
Keywords: Hta Gene; Thermus Thermophilus HB8; Homoserine Transacetylase; PHA Synthase
Abbreviations: 3HB: 3-Hydroxybutyrate; PHB: Polyhydroxybutyrate; PHA: Polyhydroxyalkanoates; PBS: Phosphate Buffered Saline; RHTA: Recombinant Homoserine Transacetylase; NAD: Nicotinamide Adenine Dinucleotide; NADP: Nicotinamide Adenine Dinucleotide Phosphate; (R)-3HBCoA: (R)-3-Hydroxybutyl-CoA Substrate; HTS: Homoserine Transsuccinylase; DMSO: Dimethyl Sulfoxide; DTNB: 5,5΄-dithio-bis(2-nitrobenzoic acid); PCR: Polymerase Chain Reaction; CBB: Coomassie Brilliant Blue; GC: Gas Chromatography; HPLC: High-Performance Liquid Chromatography; M3HB: Methyl 3‑Hydroxybutyrate; M3HV: Methyl 3‑Hydroxyvalerate; M3HE: Methyl 3‑Hydroxyhexanoate; M3HO: Methyl 3‑Hydroxyoctanoate; M3HNN: Methyl 3‑Hydroxynonanoate; M3HD: Methyl 3‑Hydroxydecanoate; M3HUD: Methyl 3‑Hydroxyundecanoate; M3HDD: Methyl 3‑Hydroxydodecanoate
Introduction
Polyhydroxyalkanoic acids (PHAs) are versatile polyesters produced by most genera of bacteria and members of the archaea as intracellular carbon and energy storage materials under nutrient-limited conditions in the presence of excess carbon source [1-3]. Since the initial discovery of polyhydroxybutyrate (PHB) in Bacillus megaterium, research has uncovered a remarkable diversity of polyhydroxyalkanoates (PHAs) with varying physicochemical properties, identified in over 90 bacterial genera. Recent studies have expanded this knowledge to include photosynthetic organisms such as cyanobacteria enhancing the potential for sustainable and innovative PHA production strategies [3]. PHAs have excellent thermoplastic and elastomeric properties, high molecular weights and in combination with their biodegradability and biocompatibility they developed into an excellent matrix for food industry, medicine, pharmacy, agriculture and chemical synthesis [4-6,2].
PHA synthases (EC 2.3.1.X) are the key enzymes for PHA biosynthesis and PHA granule assembly. They catalyze the enantio-selective polymerization of (R)-3-hydroxyacyl-CoA substrates to PHAs with the concomitant release of CoA [7,8,1]. So far, the nucleotide sequence of at least 88 PHA synthases have been obtained and the primary structures of 30 synthases are available [9,3]. PHA synthases are categorized into four classes based on their size, structure and substrate specificity [3,7,9,10], although there are known exceptions to these classes, such as PHA synthase of Aeromonas caviae [11] and synthase of Thiocapsa pfennigii [12]. The organization of PHA synthases encoding genes was of great interest and at present is fully described [3,11,13-15]. Synthases can be classified according to their primary structure into three different classes (I, II and III) [3]. An alignment of the primary sequences of PHA synthases shows an overall identity of 8-96% with 8 strictly conserved amino acids [8,10]. They belong to the α/β-hydrolase superfamily with a conserved cysteine residue as a catalytic nucleophile in a lipase box [7,16]. The N-terminus of PHA synthases is highly variable [7], though there are conserved areas within this region.
Homoserine transacetylase (HTA, EC 2.3.1.31) is a unique enzyme that catalyzes the transfer of acetate from acetyl-CoA to homoserine, in the first step of the methionine biosynthetic pathway, a pathway required for protein synthesis, nucleic acid synthesis, transmethylation reactions involving S-adenosylmethionine and the synthesis of polyamines [17,18]. The methionine biosynthetic pathway differs from one microorganism to another, regarding to the nature of the intermediates that are formed. For example, in the first step of the pathway, in E. coli, homoserine is succinylated by homoserine transsuccinylase (HTS), while in B. subtilis homoserine is acetylated. No amino acid sequence homology exists between HTS and HTA, while both enzymes use a pingpong kinetic mechanism [19]. HTA is a member of α/β-hydrolase superfamily of enzymes [20], where a catalytic triad of serine, histidine and aspartic acid is present. The catalytic nucleophile residue is found in a G-X-S/C-X-G motif [19,21-23]. Analysis of the inactivation of several HTAs by iodoacetamide (IAA) suggested that a cysteine residue acts as the nucleophile in the first half reaction to accept acetate from acetyl-CoA [18,24].
Thermus thermophilus HB8 belonging to Deinococcus- Thermus phylum, is a thermophilic bacterium with an optimum growth temperature range of 65 to 72oC [25]. It is used not only as a model for investigations on extreme thermopiles, but also as a protein pool of vigorous and thermostable enzymes that can be utilized in various biotechnological applications [26]. The biosynthesis of PHAs, in T. thermophilus grown in a mineral medium supplemented with sodium gluconate as sole carbon source has been reported previously and a cytosolic PHA synthase was purified [27].
The complete genomic sequence of T. thermophilus HB8 have been previously identified [28] and showed that it is composed of 1.85 Mbp chromosal DNA, 0.26 Mbp plasmid pTT27 and 9.32 Kbp plasmid pTT8, encoding 1973, 251 and 14 open reading frames (ORFs), respectively. However, most ORFs of the genomic sequence of T. thermophilus HB8 encode hypothetical proteins [29]. Based on various bioinformatic approaches protein function assignment was performed for all T. thermophilus ORFs but further biochemical characterization is needed to certify the assignment. TTHA0759 is one of these ORFs, that it is predicted to be a homoserine transacetylase (HTA), due to the existence of a homoserine transacetylase conserved domain. TTHA0759 is also member of the α/β-hydrolase superfamily, in which PHA synthases also belong.
The aim of our study was the identification of PHA synthase encoding gene in T. thermophillus HB8, able of producing PHAs. Searching for conserved domains of PHA synthases in T. thermophillus revealed that only TTHA0759 possess α/β- hydrolase structure and a lipase box. Thus, TTHA0759 gene (hereafter referred to as hta gene) was cloned in pET29c vector and overexpressed, after IPTG induction, as a recombinant protein with a 6xHis tag at the C-terminal. This recombinant protein was purified with metal affinity chromatography, presenting homoserine transacetylase activity, and some of its biochemical properties were studied as well. In addition, this expressed protein possesses PHA synthase activity, able to polymerize DL-3- β-hydroxybutyryl-CoA (3HBCoA) into PHB granules. The enzyme presents high affinity to PHB granules, an ability that is lost after deletion of the transmembrane domains of the protein. Finally, the in vivo production of PHAs was also determined in transformed E. coli cells with pET29c_hta, by dry cell methanolysis and GC analysis of the resulted 3-hydroxyalkanoic methyl esters.
Materials and Methods
Materials
DL-3-β-Hydroxybutyryl-CoA, acetyl-CoA, acetylphosphate, CoA, NADPH, NADP, NAD+, NADH and all other chemical compounds were purchased from Sigma-Aldrich (Chemie Gmbh, Steinheim, Germany). Restriction enzymes and other reagents required for molecular biology experiments were purchased from Takara Bio Inc. (Shiga, Japan). Plasmid vectors pET29c and pGEM-T Easy were purchased from Novagen (Darmstadt, Germany) and Promega (GmbH, Mannheim, Germany), respectively. Protino Ni-IDA columns for affinity chromatography purification were purchased from Macherey-Nagel (GmbH & Co. KG, Düren, Germany), while His-probe G18 used in immunostaining was from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA).
Cloning of hta gene
The complete coding sequence of hta gene (TTHA0759) was amplified by PCR using the genomic DNA of T. thermophilus HB8 (Takara Bio Inc.) as template. Initially, a broader region that contained hta gene was amplified in a 50 μl reaction that contained 30 pmol of each primer, 400 μM of dNTPs, 50 ng of genomic DNA, 5% v/v dimethyl sulfoxide (DMSO) and 3.0 Units of Pfu polymerase (Promega). The forward and the reverse primer for this region were 5΄-GCCTGGGTGGTCCTCCTCGG-3΄ and 5΄-CAAAGTGCTGGGCGTAGTAG-3΄, respectively. The PCR conditions were as follows: initial denaturation at 95°C for 3 min, denaturation at 95 °C for 1 min, annealing at 70°C for 1 min and polymerization at 72°C for 8 min. These steps were repeated ten times and followed by three iterations of ten cycles, in which the annealing temperature was 68°C, 63 oC and 60°C, respectively. A final elongation step was performed at 72°C for 20 min. The resulting PCR products were analyzed on 1% w/v agarose gel and cloned into pGEM-T Easy vector (Promega). JM109 competent cells (Promega) were transformed with the constructed plasmids and positive colonies were used for plasmid DNA isolation. LICOR IR2 DNA analyzer and SequeTherm sequencing kit with M13 forward and reverse universal primers were used to sequence plasmid DNA and identify hta gene.
Subsequently, the constructed plasmid was used as template DNA in a new PCR amplification where primers were designed to amplify only hta gene. The forward primer was 5΄-AAGCTTCGGGAGGAAGGCGTCCAGGATCTCCTCCAC-3΄and had a HindIII restriction site, while the reverse primer was 5΄-CATATGAGCGAGATCGCCCTCGAGGCCTGGGGG-3΄ and had an NdeI restriction site. The reaction mixture was the same as previously, while the PCR conditions were as follows: initial denaturation at 95°C for 3 min, denaturation at 95°C for 15 sec, annealing at 72°C for 15 sec and polymerization at 72°C for 45 sec. These steps were repeated thirty-five times and followed by a final elongation step at 72°C for 10 min. PCR products were cloned into pGEM-T Easy vector and transformed into JM109 competent cells. After digestion of the isolated pGEM hta plasmid with the restriction endonucleases, the fragment corresponding to HTA coding gene was ligated into the similarly digested pET29c expression vector (Novagen) to create a recombinant HTA (rHTA), containing a C-terminal histidine tag. The constructed plasmid (pET29c_hta) was finally transformed into E. coli BL21(DE3) competent cells. Preparation of competent cells, transformation and DNA isolation were performed according to Sambrook [30].
Construction of N-terminal truncated variants of HTA
For the construction of the three N-terminal truncated variants of HTA, PCR amplifications were performed using pET29c_hta as template under the conditions described above. Reverse primer was kept the same in all reactions, while the sequence of forward primers for each truncated variant, trunc_1, trunc_2 and trun_3, were 5΄-CATATGGACGAGGAAACCTTTAGAAGCC-3΄, 5 ΄ - C A T A T G C C C C T C T C C C T A G A C C C C C G - 3 ΄ , 5΄-CATATGTACCCGGAGAGGGTGAAGAAGC-3΄, respectively. PCR products were cloned into pGEM-T Easy vector and transformed into JM109 competent cells. After digestion of the isolated plasmids with the restriction endonucleases, fragments corresponding to trunc_1, trunc_2 and trunc_3 HTA coding genes were ligated into the similarly digested pET29c.
Microorganisms and growth conditions
Transformed E. coli JM109 cells with the constructed plasmid pGEM_ hta were grown at 37°C, in LB medium containing ampicilin (100 μg/ml) with vigorous agitation. E. coli BL21(DE3) harboring pET29c_hta were grown at 37°C, in LB medium containing kanamycin (50 μg/ml) with vigorous agitation. When cells reached an optical density of 0.6 at 600 nm, IPTG (1 mM) was added and the cultures were incubated at 24°C, for 3 hours.
Preparation of cell extract and purification of the recombinant protein by metal affinity chromatography
After cultivation of E. coli BL21(DE3) cells harboring pET29c_ hta as described, cells were harvested by centrifugation at 5,000xg for 15 min at 4°C. The cell pellet (2.5 g) was washed twice with PBS and suspended in 5 volumes of 50 mM NaH2 PO4 pH 8.0 containing 300 mM NaCl and 1 mg/ml lysozyme. The suspension was incubated on ice for 30 min, sonicated (5 timesx30 sec, 0.5 cycle, 50% amplitude - UP200s dr. hielscher, GmbH) and then centrifuged at 5,000xg for 20 min. The supernatant was applied to Protino Ni-IDA columns (Macherey-Nagel) equilibrated with 50 mM NaH2 PO4 pH 8.0 containing 300 mM NaCl. Elution was performed in two steps using 250 mM imidazole in the same buffer. The purified protein was dialysed against 25 mM Tris-HCl pH 8.0 and stored at -20°C.
Determination of PHB synthase activity
The PHB synthase activity was determined by a discontinuous spectrophotometric assay monitoring the release of CoA at 412 nm from the substrate DL-3-β-hydroxybutyryl-CoA, according to Müh et al. [31]. Reactions were conducted at 65°C in a final volume of 200 μl with 20 mM Tris-HCl buffer pH 8.0, 50 mM NaCl and 0.5 mM 3HB-CoA. The concentration of CoA was calculated using the extinction coefficient (412 nm) of 13,600 cm-1 M-1. One enzyme unit was defined, as the amount of enzyme required for the transformation of 1μmol substrate per min under the above-specified conditions. A continuous assay did not allow monitoring the enzyme activity, since the decomposition of 5,5΄-dithio-bis (2-nitrobenzoic acid) (DTNB), causes alterations in the absorbance at high temperature. It should be noted that in all subsequent enzyme assays controls without enzyme extract, or without substrate were used in order to monitor the non-enzymic hydrolysis occurred at the high temperature of the assay.
Determination of homoserine transacetylase activity
Homoserine transacetylase activity was determined by monitoring the change in absorbance at 232 nm due to hydrolysis or formation of the thioester bond of acetyl-CoA (ε = 4500 Μ-1) in a JENWAY 6305 UV/Vis spectrophotometer [19]. Assays were performed in 100 mM KH2 PO4 pH 8.0, at 65°C for 15 min. 0.1 mM acetyl-CoA was used as the donor and 1 mM L-homoserine as the substrate.
Dephosphorylation of rHTA
The rHTA was incubated for 30 min with various amounts of calf intestinal alkaline phosphatase bound to agarose, as previously described [27]. The mixtures were then centrifuged at 15,000xg, for 15 min and these supernatants were assayed for PHB synthase activity. As a control the same enzyme units of rHTA was incubated under the same conditions with agarose free of alkaline phosphatase.
Electrophoresis and immunoblotting
Protein electrophoresis was performed in 10% w/v polyacrylamide gels containing 0.1% w/v sodium dodecyl sulphate (SDS) as described [32]. Protein content was determined by the method of Bradford [33] using bovine serum albumin as a protein standard. Gels were stained either with Coomassie Brilliant Blue R250 (CBB), or with silver nitrate [34]. Proteins were transferred to nitrocellulose membranes following the method of Towbin [35] and immunostained with anti-His (His-probe G-18) as previously described [36].
In vitro PHB granule formation and analysis
The in vitro polymerization of 3HB-CoA to PHB granules was studied by the addition of purified rHTA. The reaction mixture contained 7 mM of 3-β-hydroxybutyryl-CoA, 100 mM of K2 HPO4 pH 8.0, and 300 μg of purified rHTA in a total volume of 1 ml. An aliquot of 20 μl of the reaction mixture was removed at various time intervals and the release of CoA was assayed with 1 mM of DTNB in 0.5 M of K2 HPO4 pH 8.0, as described [31,37]. After 1h of reaction the PHB synthesized in vitro was extracted with chloroform, the organic solvent was evaporated and the precipitated polymer was incubated in the presence of concentrated sulphuric acid at 92°C for 8h [38]. Crotonic acid formed from PHB acid-catalyzed β-elimination was analysed by HPLC on an Aminex HPX-87H ion exclusion organic acid analysis column (Bio-Rad), with the standard conditions [39], using a Shimadzu (Tokyo, Japan) High Pressure Liquid Chromatography System. Quantitation was done following comparison of peak absorbance with those of crotonic acid standards (Fluka, Buchs, Switzerland). The cPHB content of each sample was determined from the amount of crotonic acid using the established conversion rate [40]. Additionally, an infrared spectrum in the region of 4000–200 cm-1 of the synthesized polymer was obtained in KBr discs with a Perkin Elmer FT-IR 1650 spectrometer.
GC analysis of PHAs in dry cells
To determine the PHA content of transformed E. coli BL21(DE3) cells harboring pET29c_hta as well as polymer composition, 15 mg of lyophilized cells were subjected to methanolysis in the presence of 2 ml chloroform and 2 ml sulfuric acid: methanol (15:85), as previously described [41,42]. The sample was heated at 100°C for 140 min in order to obtain the corresponded 3-hydroxyalkanoic methyl esters. The monomer composition of the polymers was determined by gas chromatography (GC), with an Agilent gas chromatograph (5975c Series GC/MSD, California, USA) equipped with an HP-Innowax capillary column (30 m x 0.5 μm; J&W Scientific, California, USA) and a flame ionization detector. Two microliters of the organic phase were injected in the column. Nitrogen (1 mL/min) was used as the carrier gas. The temperature of the injector and detector were 220 and 250°C, respectively. A temperature program was used for efficient separation of the esters [50°C for 3min, temperature increase 9°C/min until 250°C, (250°C for 1 min)]. Pure standards of methyl 3-hydroxyalkanoate esters (Sigma-Aldrich Co., St Louis, MO, USA) were also analyzed under the same conditions.
Affinity with PHB granules
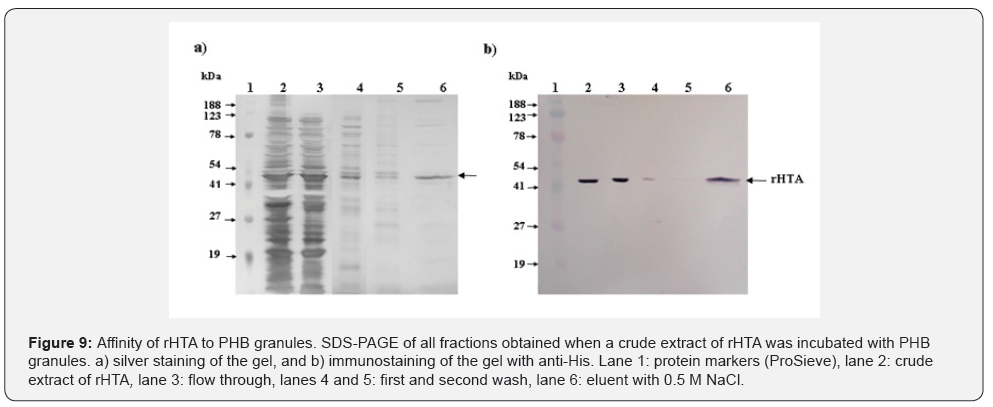
The affinity of the rHTA with PHB granules was studied by incubating 200 μg of rHTA crude extract with 4.0 mg of PHB (Sigma) in 50 mM Tris-HCl pH 8.0, at room temperature for 1h. After centrifugation (10,000xg, 15 min), PHB granules were washed three times with the same buffer and finally the bound proteins on PHB were eluted with 0.5 M NaCl in 50 mM Tris-HCl pH 8.0. The eluted proteins were dialysed against 25 mM Tris-HCl pH 8.0 and analyzed by 10% w/v SDS-PAGE. Gel was stained with silver nitrate and immunostained with anti-His respectively.
Results
We have previously reported that T. thermophilus HB8 is capable to produce PHAs, when sodium gluconate or whey were used as a carbon source [27,43]. Extending our studies we hereby attempt to identify the PHA synthase encoding gene in T. thermophilus HB8 genome and further study PHAs biosynthesis in this microorganism. A survey in the already published genomic sequence of T. thermophilus HB8 [28] was performed to identify ORFs with conserved domains of PHA synthase family. The ORF of TTHA0759 gene has an α/β-hydrolase structure and a lipase box, conserved in PHA synthase family [44]. This locus, TTHA0759, of T. thermophilus has been assigned as a putative homoserine transacetylase, based on similarities with other homoserine transacetylases [28]. In addition, when TTHA0759 protein sequence was aligned with HOMOSOATRNS (http://www.bioinf.man.ac.uk/cgi-bin/dbbrowser/prePRINTS/ searchpreprints.cgi?prints_accn=HOMOSOATRNS&display_ opts=Prints&category=None&queryform=false®expr =off&output=y), a 5-element fingerprint that provides a signature for the homoserine o-acetyltransferase proteins, all 5 motifs were identified in TTHA0759. The sequence of these motifs in Thermus aquaticus (subsp. thermophilus) is ELGGYLPEVRLRFETYGTL (Motif 1), SRRRDNAVLVFHALTGSAHLAG (Motif 2), PGRILDPALYYVVSANHLGSC YGSTGPLSL (Motif 3), EIHLDYQGEKFLRRFH (Motif 4) and AESYLVLSRAMDNHD (Motif 5).
Considering that both synthase and transacetylase possess an acyltransferase activity, we proceeded to clone and overexpress hta gene.
Cloning and overexpression of hta gene
In the cloning strategy, two pairs of primers were used in the PCR amplifications, as described in the Materials and Methods. The first pair of primers amplified a broader region that contained hta gene. This PCR product was used as DNA template in a second PCR, where primers amplified only hta gene and had specific sites for HindIII and NdeI restriction enzymes. The HindIII – NdeI fragment was finally cloned into pET29c (Novagen) expression vector.
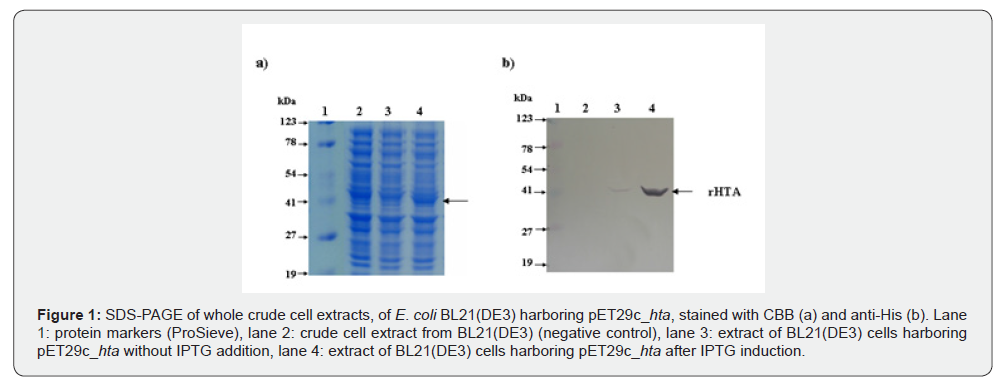
E. coli BL21(DE3) cells transformed with pET29c_hta were induced by IPTG, as described in Materials and Methods. Proteins of whole cell extracts were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with CBB (Figure 1a) and anti-His (Figure 1b). The HTA plus the C-terminal six histidine residues has a predicted molecular mass of 43 kDa. As shown in figure 1a, an intense protein band corresponding to the overexpressed rHTA was observed only in cells after IPTG induction (lane 4). The same intense band was also observed when gel was immunostained with anti-His (Figure 1b, lane 4), confirming the existence of a 6xHis tag. Before IPTG induction, a faint band was detected only with anti-His (Figure 1b, lane 3) (Figure 1).
Purification with metal affinity chromatography
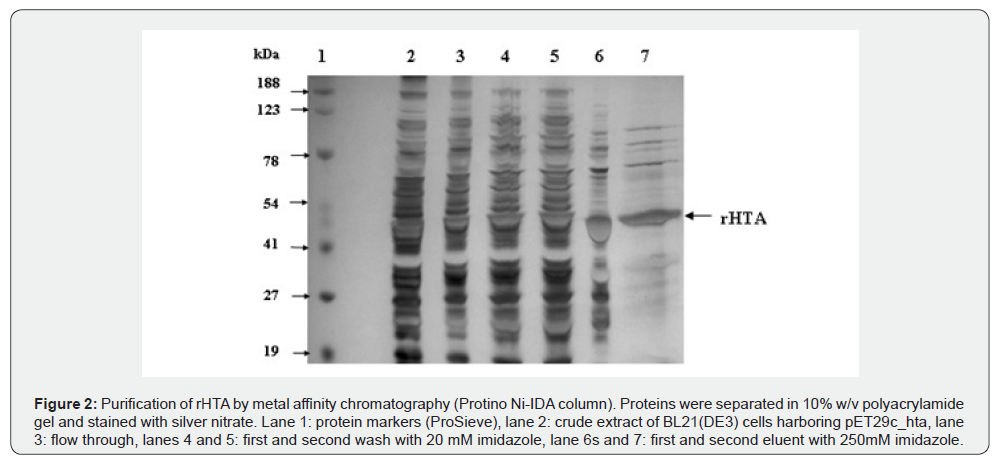
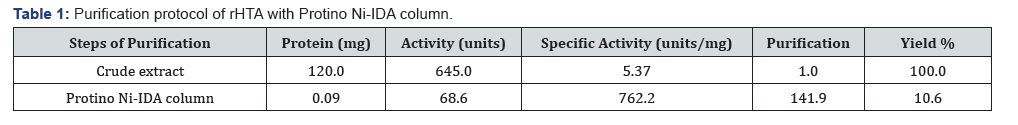
The 6xHis tag at the C-terminal enabled us to purify the protein almost to homogeneity with only one step of purification, using Protino Ni-IDA packed column. The 5,000xg supernatant from 2.5 g cells was applied on the Protino Ni-IDA column, as described in Materials and Methods. The purification achieved by this column was 142-fold with PHB synthase specific activity 762.2 units/mg and a final yield of 10.6% (Table 1). Analysis of this preparation on SDS-PAGE showed a major protein band corresponding to the rHTA (Figure 2).
Catalytic and biochemical properties of rHTA
The catalytic and biochemical characteristics of the purified enzyme were analyzed with respect to its homoserine transacetylase activity, attributed to the gene based on sequence homology. The optimal temperature of rHTA activity was about 70 °C with an optimal pH near to 7.5. The Km value as calculated from the double-reciprocal plot of velocity vs acetyl-CoA concentration was 0.11 mM.
As mentioned earlier, the rHTA of T. thermophilus possess a conserved lipase box motif and an α/β hydrolase structural domain, suggesting a potential PHA synthase activity. This function was subsequently validated through a PHB synthase assay, as described by Müh et al. [31]. Notably, the purified rHTA displayed an immediate catalytic response without a lag phase in its kinetic profile, even at low enzyme concentrations. The rate of the CoA release and consequently the incorporation of β-hydroxybutyryl unit into the polymer in vitro, was proportional to the amount of the enzyme, in the range of 2.0–60 μg of the enzyme. Above 60 μg of the purified enzyme a nonlinear dependency was observed (data obtained but not shown). The optimal temperature of rHTA regarding to PHB synthase activity was about 70°C with an optimal pH near to 7.5. The Km value for PHB synthase activity, as was estimated by the double-reciprocal plot of velocity vs 3-β-hydroxybutyryl-CoA concentration was 0.25 mM.
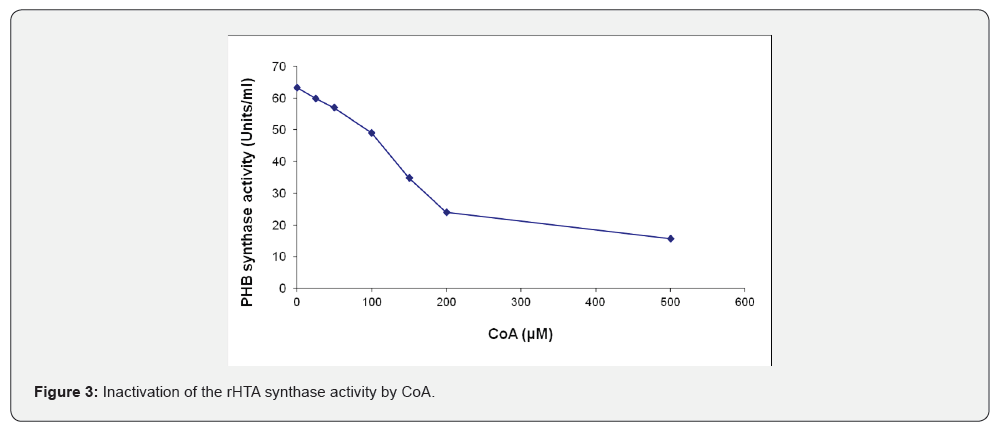
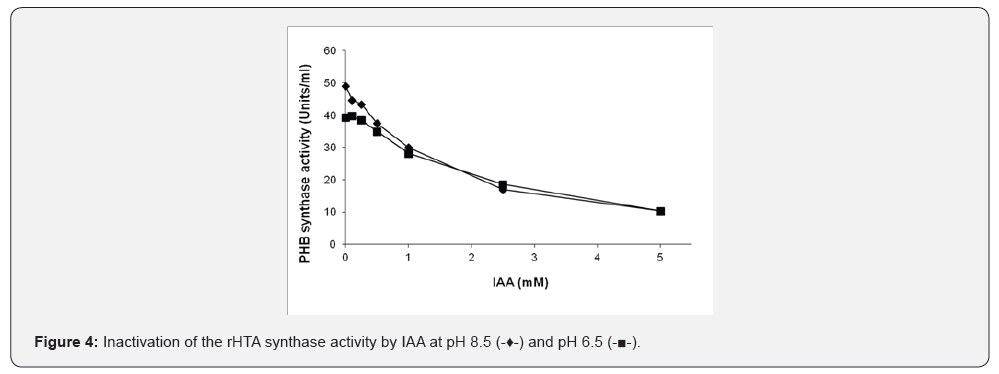
The effect of CoA on the purified rHTA from T. thermophilus was also examined. Various amounts of CoA were pre-incubated for 30 min with 60 μg of the purified rHTA prior to the determination of PHB synthase activity. The same amount of CoA was added in the control samples. PHB synthase activity was inhibited approximately 62% by the presence of 200 μM CoA and 75.2% with 500 μM of CoA, respectively (Figure 3). In addition, as shown in (Figure 4), when various concentrations of IAA were preincubated with 40 μg of the rHTA, an inhibition of enzyme activity was observed. At pH 8.5, a 65.5% inhibition was observed when 2.5 mM of IAA was used, indicating that a cysteine is involved in the catalytic site of the enzyme. As the pH of the assay buffer was decreased 6.5, a small decrease in the enzyme activity was observed. The same experiment was also conducted after 30 min of pre-incubation of rHTA with 10 mM 3-β-hydroxybutyryl-CoA, revealing the same inhibition at both pHs.
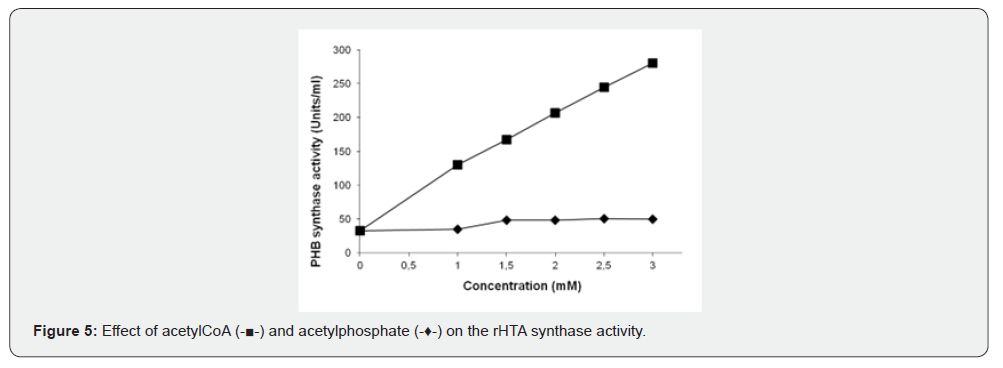
Two putative activators of PHA synthases, acetyl-CoA and acetyl phosphate were also tested for the in vitro activation of purified enzyme. Various amounts of these compounds were preincubated for 30 min with 4.0 μg of the purified enzyme prior to PHB synthase determination. The intermediate of PHA synthesis, the acetyl-CoA, at concentration of 3.0 mM, activates the purified enzyme approximately 8.5-fold. Similarly, acetyl phosphate, a putative activator of PHA synthase, activates the recombinant enzyme to 1.5-fold (Figure 5). NADH and NADPH were also used in various concentrations up to 2.5 mM without any effect on the enzyme activity. When the oxidized forms NAD+ and NADP were tested, an inhibition of approximately 25% was observed at 2.5 mM of both compounds data not shown. Experiments were performed in duplicates, and mean value is presented in all figures.
In vitro inactivation of rHTA by dephosphorylation
To test if rHTA is post-translationally modified by phosphorylation, partially purified rHTA (Protino Ni-IDA column) was incubated with increasing amounts of calf intestinal alkaline phosphatase bound to agarose and its activity was gradually decreased (data not shown), suggesting that the PHA synthase activity of rHTA is possibly regulated by a phosphorylationdephosphorylation reaction.
In vitro PHB synthesis by rHTA
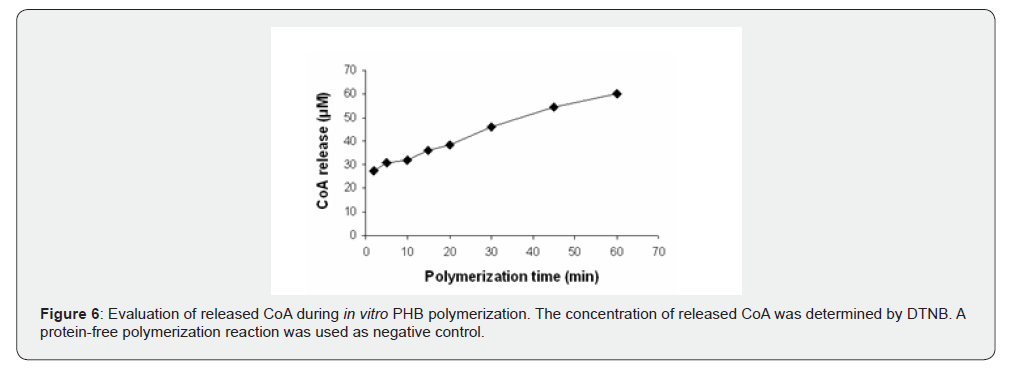
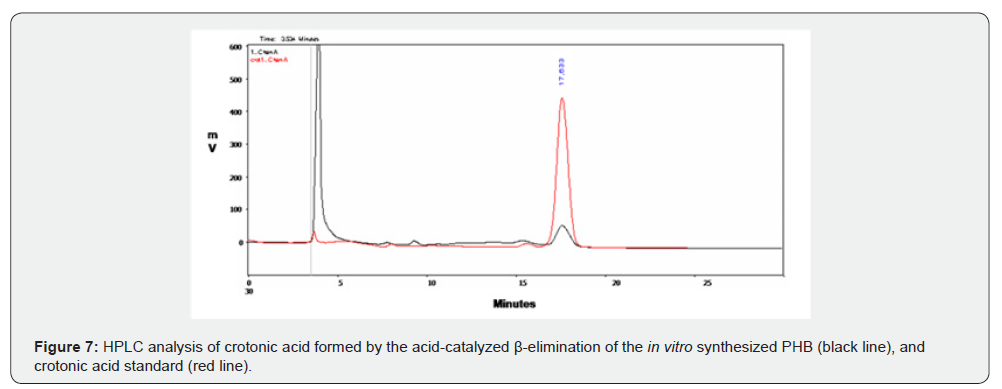
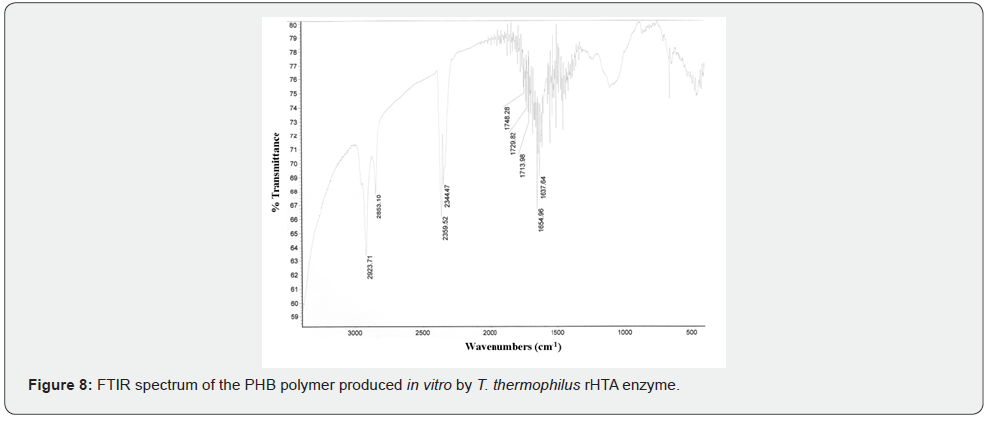
The in vitro polymerization of 3HB-CoA to PHB was performed by the purified rHTA, as described in Materials and Methods. A protein-free polymerization reaction was used as the negative control. At various time intervals, aliquots were removed from the polymerization reaction mixture and release of CoA was assayed with DTNB. As shown in (Figure 6), a linear increase of liberated CoA was observed, indicating the incorporation of β-hydroxybutyryl unit into the synthesized polymer. At the end of the reaction, synthesized PHB was extracted with chloroform and incubated with concentrated sulphuric acid as previously described [38]. The crotonic acid that was formed from PHB acid-catalyzed β-elimination, was analysed by HPLC on an Aminex HPX-87H column (Bio-Rad) as described in Materials and Methods. As shown in (Figure 7) a peak was observed at 17.633 min of retention time, corresponding to the crotonic acid. The determined cPHB content of the sample was 27.97 μM. The existence of a characteristic peak at approximately 1740 cm-1 in an IR spectrum also confirmed the in vitro PHB synthesis by purified rHTA (Figure 8).
Affinity with PHB granules
The affinity of rHTA with PHB granules was studied by incubating a crude extract of rHTA with PHB (Sigma), as described in Materials and Methods. All fractions were analyzed on 10% w/v SDS polyacrylamide gel and stained either with silver nitrate (Figure 9a), or immunostained with anti-His (Figure 9b). As shown in figure 9a lane 6, a major band corresponding to rHTA was observed in proteins eluted from PHB granules. Immunobloting with anti-His (Figure 9b, lane 6) also confirmed the existence of rHTA in the elution fraction, suggesting that rHTA has high affinity to PHB granules. It is interesting that the purification performed with PHB granules was even better than the purification achieved with Protino Ni-IDA (Figure 9).
Deletion of the transmebrane domains of rHTA
To study whether the transmembrane domains of rHTA are responsible for its binding to PHB granules, three truncated variants of rHTA were constructed and their affinity with PHB was determined as described with the full-length protein.
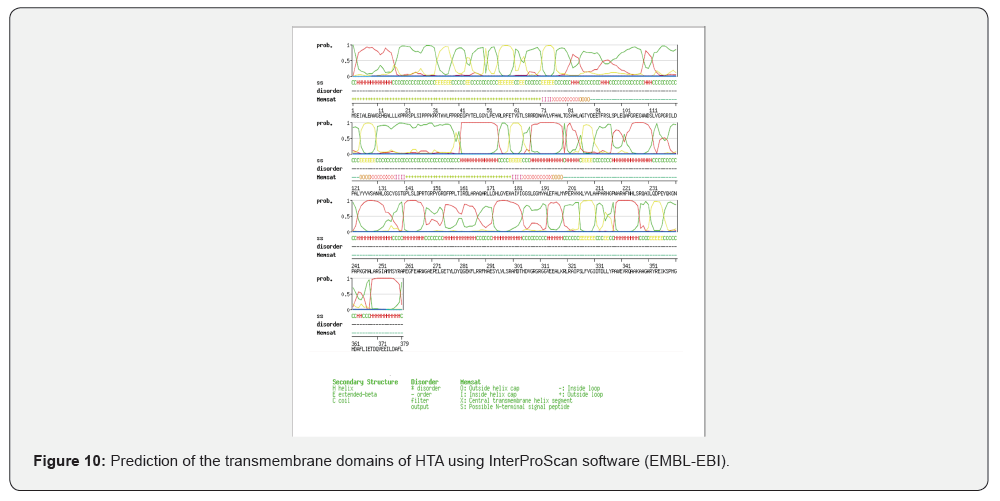
InterProScan software package (EMBL-EBI) was used to determine the transmembrane domains of rHTA as shown in (Figure 10). Three transmembrane domains were identified at 75-84, 128-136 and 184-194 aa of rHTA. Deletion of these domains was achieved by PCR using primers downstream of the corresponding coding sequences, as described in Material and Methods. The truncated variants of rHTA were overexpressed after IPTG induction and the crude extracts of each variant was incubated with PHB granules under the same conditions used for the full length of rHTA. Analysis of all fractions with SDS-PAGE electrophoresis followed by staining with silver nitrate revealed that the truncated variants of rHTA have no affinity with PHB.
PHA production by recombinant E. coli
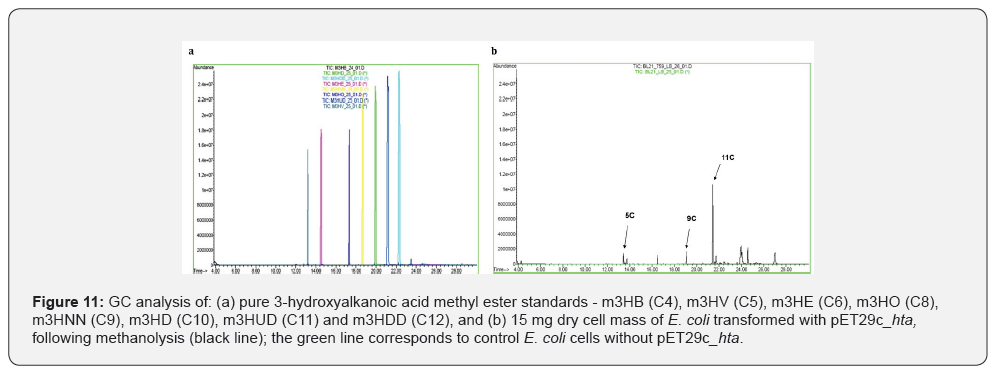
To determine the PHA content of transformed E. coli BL21(DE3) cells harboring pET29c_hta as well as polymer composition, 15 mg of lyophilized cells were subjected to methanolysis as previously described [41,42]. The resulted 3-hydroxyalkanoic methyl esters were determined by GC analysis, while various 3-hydroxyalkanoic acid methyl ester standards were also analyzed under the exact same conditions. The standards employed in the analysis were methyl 3‑hydroxybutyrate (m3HB), methyl 3‑hydroxyvalerate (m3HV), methyl 3‑hydroxyhexanoate (m3HE), methyl 3‑hydroxyoctanoate (m3HO), methyl 3‑hydroxynonanoate (m3HNN), methyl 3‑hydroxydecanoate (m3HD), methyl 3‑hydroxyundecanoate (m3HUD) and methyl 3‑hydroxydodecanoate (m3HDD). The retention time for each standard is presented in Figure 11a. As shown in figure 11b (black line), the GC analysis of the polymer produced by the transformed E. coli cells revealed the presence of pentanoic, nonanoic, and undecanoic acid monomers, as determined by the retention time of the corresponding peaks. GC analysis of the methanolysis products from E. coli cells not carrying the pET29c_hta revealed no detectable peaks, indicating the absence of PHA accumulation (Figure 11).
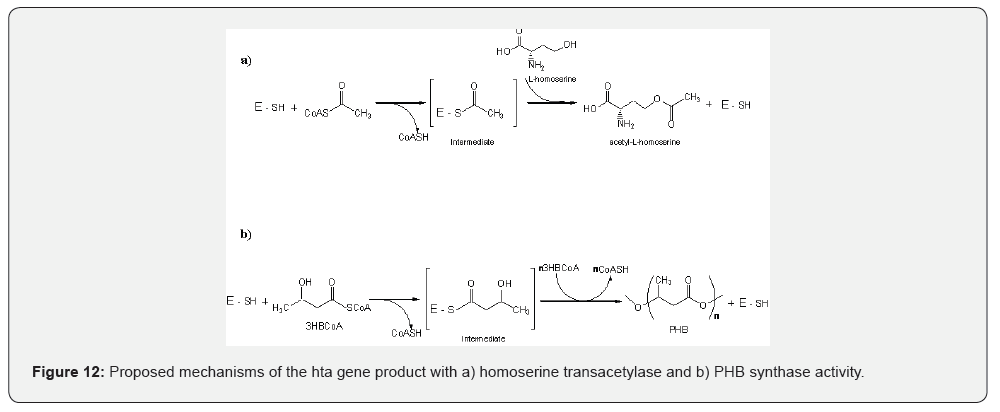
Discussion
This study aimed to identify the gene encoding polyhydroxyalkanoate (PHA) synthase in T. thermophilus HB8, a thermophilic bacterium previously characterized as a PHAaccumulating microorganism [27]. Our group has previously purified and biochemically characterized two PHA biosynthetic enzymes, a PHA synthase and a β-ketothiolase [45,27]. A genome-wide survey for loci exhibiting an α/β-hydrolase fold and containing the conserved G-X-S/C-X-G lipase box motif in T. thermophilus HB8 identified the TTHA0759 locus. Although bioinformatic analysis previously annotated this locus as encoding a homoserine transacetylase [28], its structural features suggested potential PHA synthase activity. Homoserine transacetylases are members of the α/β-hydrolase superfamily and it was found that the G-G-S-X-G-G sequence is conserved among HTAs. There are also three conserved serine, two histidine and six aspartic acid residues [19].
Previous studies on TmHTA [18] and Haemophilus influenzae HTA [19] demonstrated that acetyl-CoA is the favoured acyldonor, while D-homoserine and L-homoserine are the best acyl acceptors. Acyl donors such as, butyryl-CoA and β-hydroxybutyryl- CoA displayed minimal activity with Tm HTA, while Hi HTA is able to accommodate four carbon acyl side chains, such as butyryl-CoA [19]. When purified rHTA transacetylase activity was assayed using acetyl-CoA and L-homoserine as substrates, the Km for acetyl-CoA was 110 μM, a value very close to 130 ± 20 μM of TmHTA and 140 ± 10 μM of Hi HTA. The optimum temperature of rHTA was around 70°C as expected for an enzyme expressed by a thermophilic bacterium.
To confirm that HTA of T. thermophilus HB8 also functions as a PHA synthase, purified rHTA was subjected to a standard PHA synthase assay. The enzyme exhibited significant synthase activity, and its kinetic properties were compared with those of other known PHA synthases. The Km value of the purified rHTA, as calculated from the double-reciprocal plot of reaction velocity versus 3-β-hydroxybutyryl-CoA concentration, was 0.25 mM, identical to the Km value reported for the native PHA synthase of T. thermophilus [27]. Notably, the synthase activity was markedly reduced following dephosphorylation of the purified rHTA by an alkaline phosphatase, suggesting that phosphorylation is a crucial post-translational modification for its catalytic function. The same loss of activity upon dephosphorylation has also been observed in the native T. thermophilus PHA synthase and in other PHA or PHB synthases [46,47].
Studies on Thermotoga maritima HTA showed that Tm HTA contains the putative catalytic cysteine residue of the homoserine transsuccinylase (HTS) family [18] suggesting that in some members of HTA family the serine residue of the lipase box is replaced by cysteine. Inactivation of rHTA activity was studied with IAA indicating that a cysteine residue is involved in the catalytic site of this enzyme and that T. thermophilus HTA is another member of the HTA family that has a catalytic cysteine residue instead of serine. Inactivation by IAA was also revealed in the native PHA synthase of T. thermophilus [27] and it is in accordance with all PHA synthases, where the residues cysteine, aspartate and histidine were shown to be essential for covalent catalysis. Cysteine is the catalytic nucleophile, histidine activates cysteine and aspartate functions as a general base to activate the 3-hydroxyl group of HBCoA, or a covalently bound HB for nucleophilic attack [48]. A proposed mechanism for the homoserine transacetylase activity and synthase activity of the rHTA is given in in figure 12. For both reactions an intermediate with the substrate is formed, attached to the thiol group of cysteine residue, and CoA is removed. In the transacetylase reaction, the acyl moiety is transferred to L-homoserine, while in the synthase reaction the hydroxyacyl moiety is transferred to another hydroxyacyl moiety thus forming the polymer chain.
To evaluate the synthase activity of rHTA in comparison with the native PHA synthase from T. thermophilus and with other known PHA synthases, various compounds recognized as PHA synthase activators or inhibitors were tested. Acetyl-CoA, an intermediate of PHA biosynthesis, and acetylphosphate, a putative activator of PHA synthase, at a concentration of 2.5 mM enhanced the activity of purified rHTA by approximately 8.5-fold and 1.5-fold, respectively, consistent with the activation patterns observed in other PHA synthases, including the native synthase from T. thermophilus [27]. NADH and NADPH were also tested at concentration up to 2.5 mM, without any effect on the enzyme activity, while NAD+ and NADP at the same concentration led to a ~25% inhibition of enzyme activity. Notably, previous studies reported that NADH and NADPH do not alter synthase activity. However, the effect of NAD+ and NADP on synthase activity was more profound resulting in the total loss of enzyme activity.
The most convincing evidence that rHTA of T. thermophilus HB8 exhibits PHA synthase activity includes (i) its ability to catalyze the in vitro polymerization of 3-β-hydroxybutyryl-CoA into PHB, (ii) its strong affinity for PHB granules, and (iii) the production of PHAs in E. coli cells transformed with pET29c_hta, observed only upon IPTG induction, as confirmed by GC analysis of methanolysis products from the dry cell mass (Figure 11b).
In the first case, the formation of PHB granules was confirmed through multiple analytical approaches: (i) CoA release was monitored, indicating the incorporation of β-hydroxybutyryl unit into the produced polymer, (ii) the synthesized polymer was subjected to concentrated sulfuric acid treatment at 92°C for 8 hours, resulting in the formation of crotonic acid, which was subsequently detected by HPLC; and (iii) a characteristic peak at 1740 cm-1 was appeared in the IR spectrum, consistent with previously reported data [38,49,50]. In vitro polymerization of 3HBCoA into PHB was demonstrated by PHA synthases from various microorganism, such as Chromatium vinosum [49] and Alcaligenes eutrophus [50]. Parameters that affect PHB formation were studied and provided useful information required to optimize the PHA production process in bacteria and recombinant microorganisms.
Previous studies have shown that PHA synthase exhibits strong affinity for PHB granules and is covalently anchored to granule surface [9]. Moreover, the N-terminal region of PHA synthases has been successfully employed as a tag for the purification of recombinant proteins using PHB granules as a matrix [51-53]. In our study, the affinity of crude rHTA for PHB was assessed as described in the Results section. Elution of bound proteins with 0.5 M NaCl followed by SDS-PAGE analysis revealed a prominent band corresponding to rHTA, indicating its high affinity for PHB granules. Notably, the purification achieved using PHB granules exceeded that obtained with the Protino Ni-IDA column.
In PHA-accumulating cells, PHA granules are typically surrounded by a phospholipid monolayer embedded with or associated to various proteins, notably PHA synthase [54]. To investigate the potential interaction between PHB synthase and the cytoplasmic membrane, R. eutropha PHB-4 cells (deficient in endogenous PHB synthase) were transformed with a plasmid encoding a phaC-eGFP fusion protein. Fluorescence microscopy revealed significant fluorescence signals along with the cytoplasmic membrane and occasionally minute signals at the cell periphery in the form of very tiny granular spots [54]. These observations suggest that the fusion protein may be associated with the membrane, potentially reflecting the native localization behavior of PHB synthase. In our study, deletion of the transmembrane domains resulted in the loss of PHB-binding activity. This suggests that the transmembrane domains of T. thermophilus HTA play a crucial role in anchoring or orienting HTA in a way that facilitates PHB interaction. It is possible that these domains contribute to the structural conformation necessary for binding, or they may help localize HTA to membrane regions rich in PHB precursors.
Finally, the detection of 3-hydroxyalkanoic acid methyl esters via GC analysis of the methanolysis product of E. coli cells transformed with pET29c_hta confirms the biosynthesis of PHAs. Specifically, the presence of peaks corresponding to pentanoic, nonanoic, and undecanoic acid monomers suggests that the recombinant strain is capable of producing medium-chain-length PHAs. The lack of detectable peaks in the GC analysis of the methanolysis of E. coli control cells indicates that the observed polymer production is not due to endogenous metabolic activity of E. coli, instead it represents a direct consequence of the genetic construct that was introduced.
In conclusion, our findings revealed that hta gene of T. thermophilus HB8 encodes a protein with homoserine transacetylase activity that also functions as a PHA synthase. In vitro PHB biosynthesis was also established with rHTA of T. thermophilus. Further studies are needed to optimize in vitro PHB formation and to elucidate PHA biosynthetic pathway in T. thermophilus HB8.
Author’s Contribution
RMP carried out all the experimentation, acquisition and analysis of data, drafting and revising of the manuscript. DAK conceived, designed, provided funding and supervised the study, and revised the manuscript. All authors have read and approved the final manuscript.
Acknowledgments
This work was carried out in the framework of the IPproject ‘Sustainable Microbial and Biocatalytic Production of Advanced Functional Materials’ (BIOPRODUCTION/NMP-2- CT-2007-026515) funded by the European Commission; the contents of the publication reflect only the author’s view.
References
- Zher Neoh S, Fey Chek M, Tiang Tan H, Linares-Pastén JA, Nandakumar A, et al. (2022) Polyhydroxyalkanoate synthase (PhaC): The key enzyme for biopolyester synthesis. Curr Res Biotechnol 4: 87-101.
- Sudesh K, Abe H, Doi Y (2000) Synthesis, structure and properties of polyhydroxyalkanoates: Biological polyesters. Prog Polym Sci 25(10): 1503-1555.
- Giourieva V, Papi RM, Pantazaki AA (2018) Polyhydroxyalkanoates: New Browsing the PHA Biosynthesis Insights in Native and Recombinant Strains. In: Williams H, Kelly P (eds), Polyhydroxyalkanoates: Biosynthesis, Chemical Structures and Applications. Nova Science Publishers Inc. Hauppauge NY 11788 USA pp. 71-110.
- Halevas EG, Andriotis EG, Papi RM, Pantazaki AA (2018) Polyhydroxyalkanoates: An Ideal Polymeric Material in Food Packaging. In: Williams H, Kelly P (eds), Polyhydroxyalkanoates: Biosynthesis, Chemical Structures and Applications. Nova Science Publishers Inc., Hauppauge NY 11788 USA pp. 287-305.
- Anderson AJ, Dawes EA (1990) Occurrence, metabolism, metabolic role and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Mol Biol Rev 54(4): 450-472.
- Williams SF, Martin DP, Horowitz DH (1999) PHA applications: Addressing the price performance issue: I. Tissue engineering. Int J Biol Macromol 25(1-3): 111-121.
- Rehm BHA (2003) Polyester synthases: Natural catalysts for plastics. Biochem J 376: 15-33.
- Rehm BHA (2006) Genetics and biochemistry of polyhydroxyalkanoate granule self-assembly: The key role of polyester synthases. Biotechnol Lett 28(4): 207-213.
- Grage K, Jahns AC, Parlane N, Palanisamy R, Rasiah IA, et al. (2009) Bacterial polyhydroxyalkanoate granules: Biogenesis, structure, and potential use as nano-/micro-beads in biotechnological and biomedical applications. Biomacromolecules 10(4): 660-669.
- Nomura CT, Taguchi S (2007) Pha synthase engineering toward superbiocatalysts for custom-made biopolymers. Appl Microbiol Biotechnol 73(5): 969-979.
- Fukui T, Doi Y (1997) Cloning and analysis of the poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) biosynthesis genes of Aeromonas caviae. J Bacteriol 179(15): 4821-4830.
- Liebergesell M, Mayer J, Steinbüchel A (1993) Analysis of polyhydroxyalkanoic acid biosynthetic genes of anoxygenic phototrophic bacteria reveals synthesis of a polyester exhibiting an unusual composition. Appl Microbiol Biotechnol 40: 292-300.
- Pieper U, Steinbüchel A (1992) Identification, cloning and sequence analysis of the poly (3-hydroxyalkanoic acid) synthase gene of the gram-positive bacterium Rhodococcus ruber. FEMS Microbiol Letts 75(1): 73-80.
- Matsusaki H, Manji S, Taguchi S, Kato M, Fukui T, et al. (1998) Cloning and molecular analysis of the poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyalkanoate) biosynthesis genes in Pseudomonas sp. Strain 61-3 J Bacteriol 180(24): 6459-6467.
- Liebergesell M, Rehalkar S, Steinbüchel A (2000) Analysis of the Thiocapsa pfennigii polyhydroxyalkanoate synthase: Subcloning, molecular characterization and generation of hybrid synthases with the corresponding Chromatium vinosum enzyme. Appl Microbiol Biotechnol 54(2): 186-194.
- Jia Y, Yuan W, Wodzinska J, Park C, Sinskey AJ, et al. (2001) Mechanistic studies on class I polyhydroxybutyrate (PHB) synthase from Ralstonia eutropha: Class I and class III share a similar catalytic mechanism. Biochemistry 40(4): 1011-1019.
- Saint-Girons I, Parsot C, Zakin MM, Bârzu O, Cohen GN (1988) Methionine biosynthesis in Enterobacteriaceae: biochemical, regulatory, and evolutionary aspects. CRC Crit Rev Biochem. 23(Suppl 1): S1-42.
- Goudarzi M, Born TL (2006) Purification and characterization of Thermotoga maritima homoserine transsuccinylase indicates it is a transacetylase. Extremophiles 10(5): 469-478.
- Born TL, Franklin M, Blanchard JS (2000) Enzyme-catalyzed acylation of homoserine: Mechanistic characterization of the Haemophilus influenzae met2-encoded homoserine transacetylase. Biochemistry 39(29): 8556-8564.
- Nazi I, Wright GD (2005) Catalytic mechanism of fungal homoserine transacetylase. Biochemistry 44(41): 13560-13566.
- Hemila H, Koivula TT, Palva I (1994) Hormone-sensitive lipase is closely related to several bacterial proteins and distantly related to acetylcholinesterase and lipoprotein lipase: Identification of a superfamily of esterases and lipases. Biochim Biophys Acta 1210(2): 249-253.
- Brumlik MJ, Buckley JT (1996) Identification of the catalytic triad of the lipase/acyltransferase from Aeromonas hydrophila. J Bacteriol 178(7): 2060-2064.
- Schrag JD, Cygler M (1997) Lipases and alpha/beta hydrolase fold. Methods Enzymol 284: 85-107.
- Ziegler K, Yusupov M, Bishop B, Born TL (2007) Substrate analysis of homoserine acyltransferase from Bacillus cereus. Biochem Biophys Res Commun 361(2): 510-515.
- Brock TD (1969) In: Krieg NR, Holt JG (eds) Bergey's Manual of Systematic Bacteriology Williams & Wilkins pp. 333-339.
- Pantazaki AA, Pritsa AA, Kyriakidis DA (2002) Biotechnologically relevant enzymes from Thermus thermophilus. Appl Microbiol Biotechnol 58(1): 1-12.
- Pantazaki AA, Tambaka MG, Langlois V, Guerin P, Kyriakidis DA (2003) Polyhydroxyalkanoate (PHA) biosynthesis in Thermus thermophilus: Purification and biochemical properties of PHA synthase. Mol Cell Biochem 254(1-2): 173-183
- Henne A, Bruggemann H, Raasch C, Wiezer A, Hartsch T, et al. (2004) The genome sequence of the extreme thermophile Thermus thermophilus. Nature Biotechnology 22(5): 547-553.
- Lioliou EE, Pantazaki AA, Kyriakidis DA (2004) Thermus thermophilus genome analysis: Benefits and implications. Microb Cell Fact 3: 5.
- Sambrook J, Fritsch E, Maniatis T (1989) Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press Cold Spring Harbor.
- Müh U, Sinskey A, Kirby DP, Lane WS, Stubbe J (1999) PHA synthase from Chromatium vinosum: Cysteine 149 is involved in covalent catalysis. Biochemistry 38(2): 826-837.
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259): 680-685.
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Blum H, Beir H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gel. Electrophoresis 8(2): 93-99.
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci 76(9): 4350-4354.
- Blake MS, Johnston KH, Russels-Jones GJ, Gotschlich EC (1984) A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on western blots. Anal Biochem 136(1): 175-179.
- Ellman GL (1959) Tissue sulfhydryl groups. Arch Biochem Biophys 82(1) :70-77.
- Theodorou MC, Panagiotidis CA, Panagiotidis CH, Pantazaki AA, Kyriakidis DA (2006) Involvement of the AtoS-AtoC signal transduction system in poly-(R)-3-hydroxybutyrate biosynthesis in Escherichia coli. Biochim Biophys Acta 1760(6): 896-906.
- Huang R, Reusch RN (1996) Poly(3-hydroxybutyrate) is associated with specific proteins in the cytoplasm and membranes of Escherichia coli. J Biol Chem 271(36): 22196-22202.
- Taguchi S, Maehara A, Takase K, Nakahara M, Nakamura H, et al. (2001) Analysis of mutational effects of a polyhydroxybutyrate (PHB) polymerase on bacterial PHB accumulation using an in vivo assay system. FEMS Microbiol Lett 198(1): 65-71.
- Braunegg G, Sonnleitner B, Lafferty RA (1978) Rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Eur J Appl Microbiol Biotechnol 6: 29-37.
- Tappel RC, Wang Q, Nomura CT (2012) Precise control of repeating unit composition in biodegradable poly(3-hydroxyalkanoate) polymers synthesized by Escherichia coli. J Biosci Bioeng 113(4): 480-486.
- Pantazaki AA, Papaneophytou CP, Pritsa AG, Liakopoulou-Kyriakides M, Kyriakidis DA (2009) Production of polyhydroxyalkanoates from whey by Thermus thermophilus HB8. Process Biochemistry 44(8): 847-853.
- Stubbe J, Tian J (2003) Polyhydroxyalkanoate (PHA) homeostasis: The role of the PHA synthase. Nat Prod Rep 20(5): 445-457.
- Pantazaki AA, Ioannou AK, Kyriakidis DA (2005) A thermostable β-ketothiolase of polyhydroxyalkanoates (PHAs) in Thermus thermophilus: Purification and biochemical properties. Molecular and Cellular Biochemistry 269(1-2): 27-36.
- Gerngross TU, Snell KD, Peoples OP, Csuhai E, Masamune S, et al. (1994) Overexpression and purification of the soluble polyhydroxyalkanoates synthase from Alcaligenes eutrophus: Evidence for a required posttranslational modification for catalytic activity. Biochemistry 33(31): 9311-9320.
- Miyake M, Kataoka K, Shirai M, Asada Y (1997) Control of poly-β-hydroxybutyrate synthase mediated by acetyl phosphate in cyanobacteria. J Bacteriol 179(16): 5009-5013.
- Jia Y, Kappock T, Frick T, Sinskey AJ, Stubbe J (2000) Lipases provide a new mechanistic model for polyhydroxybutyrate (PHB) synthases: Characterization of the functional residues in Chromatium vinosum PHB synthase. Biochemistry 39(14): 3927-3936.
- Jossek R, Reichelt R, Steinbüchel A (1998) In vitro biosynthesis of poly (3-hydroxybutyric acid) by using purified poly (hydroxyalkanoic acid) synthase of Chromatium vinosum. Appl Microbiol Biotechnol 49: 258-266.
- Gerngross TU, Martin DP (1995) Enzyme-catalyzed synthesis of poly[(R)-(-)-3-hydroxybutyrate]: Formation of macroscopic granules in vitro. Proc Natl Acad Sci 92(14): 6279-6283.
- Jahns AC, Haverkamp RG, Rehm BHA (2008) Multifunctional inorganic-binding beads self-assembled inside engineered bacteria. Bioconjugate Chem 19(10): 2072-2080.
- Grage K, Rehm BHA (2008) In vivo production of scfv-displaying biopolymer beads using a self-assembly-promoting fusion partner. Bioconjugate Chem 19(1): 254-262.
- Peters V, Rehm BHA (2008) Protein engineering of streptavidin for in vivo assembly of streptavidin beads. J Biotechnol 134(3-4): 266-274.
- Jendrossek D (2009) Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J Bacteriol 191(10): 3195-3202.






























