Technical Sheet of Relative Expression of the TIF11 and YJL144W Genes Coding for Hydrophilins that Contribute to the Freeze-Drying Tolerance of Yeasts Strains (Saccharomyces Cereviviae and Candida Tropicalis) Isolated from Traditional Sorghum Beer to Côte d’Ivoire
Coulibaly Wahauwouele Hermann1*, Boli Zamblé Bi Irié Abel1, Bouatenin Koffi Maïzan Jean-Paul1, Kouamé Hanzi Karen1, WAM Alloue-Boraud1, Marlène Cot2 and Djè Koffi Marcellin1
1Department of Food Science and Technology, Biotechnology and Food Microbiology Laboratory, University Nangui Abrogoua, Abidjan, Côte d’Ivoire
2CRT/CRITT Bio-Industries, INSA Toulouse 135 avenue de Rangueil 31077 Toulouse CEDEX 04, France
Submission: March 09, 2019; Published: May 03, 2019
*Corresponding author: Coulibaly Wahauwouele Hermann, Department of Food Science and Technology, Biotechnology and Food Microbiology Laboratory, University Nangui Abrogoua, Abidjan, Côte d’Ivoire
How to cite this article: Coulibaly Wahauwouele Hermann, Boli Zamblé Bi Irié Abel, et al. Technical Sheet of Relative Expression of the TIF11 and YJL144W Genes Coding for Hydrophilins that Contribute to the Freeze-Drying Tolerance of Yeasts Strains (Saccharomyces Cereviviae and Candida Tropicalis) Isolated from Traditional Sorghum Beer to Côte d’Ivoire. Adv Biotech & Micro. 2019; 13(5): 555873. DOI: 10.19080/AIBM.2019.13.555873
Abstract
Hydrophilins are a group of proteins that are present in all organisms and that have been defined as being highly hydrophilic and rich in glycine. They are assumed to play important roles in cellular dehydration tolerance. There are 12 genes in the yeast Saccharomyces cerevisiae that encode hydrophilins and most of these genes are stress responsive. However, the functional role of yeast hydrophilins, especially in desiccation and freeze-drying tolerance, is largely unknown. Here, we selected two genes coding for hydrophilins for further analysis. The TIF11 gene was more expressed than the YJL144W gene and this for all the strains of the two yeast species studied. For S. cerevisiae strains, the high expressions levels of TIF11 and YJL144W were obtained with D12-3 and E4-4 strains with respectively 5.56 ± 0.89 and 3.37 ± 0.48. For C. tropicalis strains, the high expressions levels were 2.95 ± 0.12 for F0-5 strains and 2.23 ± 0.70 for C8-10 strains respectively for TIFF11 and YJL144W. For C. tropicalis strains, the statistical analyzes of the relative expression levels from the Tukey test revealed no difference between the strains for the 2 genes (P> 0.05).
Keywords: Saccharomyces cerevisiae; Candida tropicalis; Hydrophilins; TIF11 and YJL144W genes; Freeze-drying; Traditional sorghum beer
Introduction
Hydrophilins are a group of highly hydrophilic and glycine-rich proteins, which are hypothesized to play a role in cellular dehydration tolerance. The yeast genome includes 12 genes encoding hydrophilins and most of them are stress responsive [1]. They are assumed to play important roles in cellular dehydration tolerance. There are 12 genes in the yeast that encode hydrophilins and most of these genes are stress responsive [1]. In many studies, the microorganisms were used for the production of freeze-dried starter culture without known to capacities to resist to dehydration stress. The viability was enhanced by adding many protective compounds such as disaccharides, polyols, monosaccharides, skim milk, and other organic molecules [2]. According to the work of Berny and Hennebert [3], by using skim milk as a support material in combination with two compounds between honey, sodium glutamate, trehalose or raffinose, the viability of Saccharomyces cerevisiae cells increased from 30% to 96-98%. Abadias et al. [4]. reported a survival rate of 28.9% for Candida sake when 10% skim milk was used. Zhao & Zhang [5] obtained the highest viability (53.6%) after freeze-drying of Oenococcus oeni H-2 by using 2.5% sodium glutamate. But the adding many protectives compounds can be of economic disadvantage in developing countries.
To date the study of yeasts intrinsic capacities to resist to dehydration and stress tolerance for the freeze-drying starter culture production for traditional sorghum remains untapped. In this study, we aimed to select strains of Saccharomyces cerevisiae and Candida tropicalis based on evaluate the expression level of the hydrophillins encoding genes TIF 11 and YJL144W to express encoding for hydrophilins.
Materials and Methods
Yeasts Strains
The yeast strains used in this study were Candida tropicalis (C0-7; F0-5; C8-10) and Saccharomyces cerevisiae (D12-3; D12- 10; E4-4; A12-1; C8-5; F12-7), preserved in the culture collection of the Department of Food Technology (University of Nangui- Abrogoua, Abidjan, Côte d’Ivoire). These strains were originally isolated from traditional sorghum beer from the district of Abidjan (Southern Côte d’Ivoire) and were thereafter identified through Polymerase Chain Reaction-Restriction Fragment Length polymorphism (PCR-RFLP) analysis of the Internal Transcribed Spacer (ITS) region and sequencing of D1/D2 domains of the 26S rRNA gene [6]. Yeast strains were maintained routinely at -20 °C in 20% glycerol.
Relative Expression of the TIFF11 and YJL144W Genes through Quantitative Real-time PCR (qRT-PCR)
Yeast strains, growth conditions and sampling
The Saccharomyces cerevisiae strains (D12-3; D12-10; E4-4; A12-1; C8-5; F12-7) and Candida tropicalis strains (C0-7; F0-5; C8- 10) were grown at 30 °C in a synthetic minimal medium containing 0.17% (w/v) yeast nitrogen base (DIFCO), 0.5% (w/v) ammonium sulfate and 2% (w/v) galactose (YNGal) or glucose (YNGlu). Prototroph strains were used in order to prevent amino acid complementation of the medium. The pH was adjusted to 5.0 with succinic acid and sodium hydroxide. Cell growth was monitored through periodical OD600nm measurements over at least 10 days. Replicate culture flasks were generated independently at different times from distinct inocula. Yeast samples for real-time PCR analysis (approx. 108 cells) were immediately centrifuged, then cell pellets were flash-frozen in liquid nitrogen and stored at -80 °C until used for RNA extractions
Total RNA extraction
Frozen cells were mechanically disrupted using a ball mill (Mikro-Dismembrator S; B. Braun Biotech International). Total RNA was extracted using the RNeasy mini kit (Qiagen) according to the manufacturer’s instruction. The purity and concentration of the extracted RNA were assessed spectrophotometrically using the ND1000 UV-visible light spectrophotometer (NanoDrop Technologies) and its integrity was checked with the Bioanalyzer 2100 with the RNA 6000 Nano LabChip kit (Agilent).
Quantitative RT-PCR
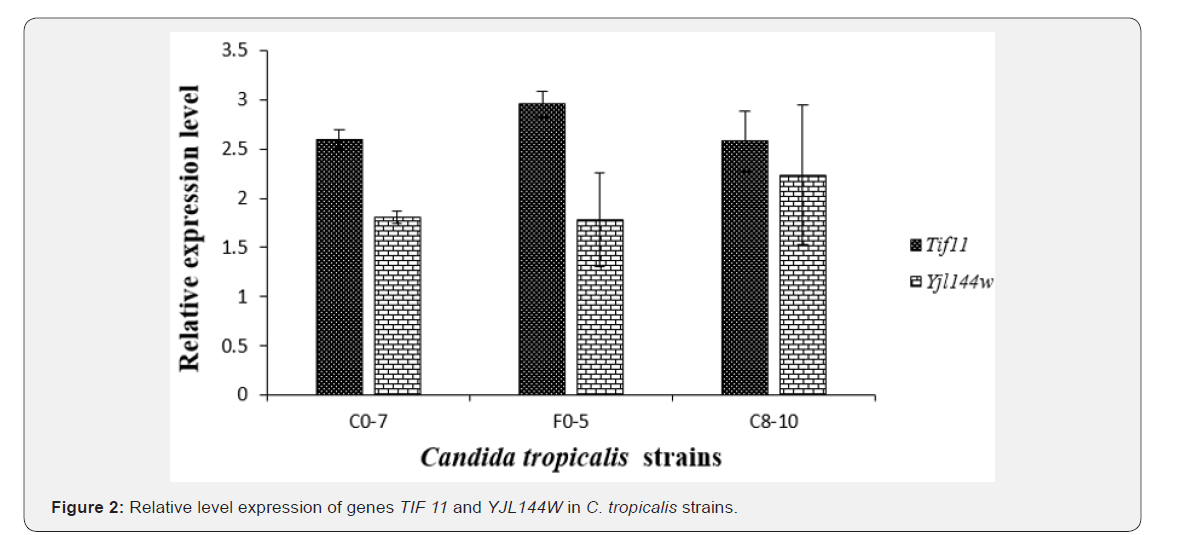
Oligonucleotides for real-time PCR (Table 1) were designed using Beacon Designer 2.0 software (PREMIER Biosoft International), which included a BLAST analysis against Saccharomyces cerevisiae and Candida tropicalis Genome sequence for specificity confidence, and analysis using the Mfold server to avoid positioning on unfavorable secondary structures.
One microgram of total RNA was reverse transcribed into cDNA in a 20 μL reaction mixture using the iScript cDNA synthesis kit (Bio-Rad). This experience was repeated three times. The cDNA levels were then analyzed using the MyIQ real-time PCR system from Bio-Rad. Each sample was tested in duplicate in a 96- well plate (Bio-Rad, CA). The reaction mix (25 μL final volume) consisted of 12.5 μL of iQ SYBR Green Supermix (BioRad), 2.5 μL of each primer (250 nM final concentration), 2.5 μL of H2O, and 5 μL of a 1/10 dilution of the cDNA preparation. The absence of genomic DNA in RNA samples was checked by real-time PCR before cDNA synthesis (minus RT control). A blank (No Template Control) was also incorporated in each assay. The thermocycling program consisted of one hold at 95 °C for 4 min, followed by 40 cycles of 10 s at 95 °C and 45 s at 56 °C. After completion of these cycles, melting-curve data were then collected to verify PCR specificity, contamination and the absence of primer dimers.
The PCR efficiency of each primer pair (Eff) was evaluated by the dilution series method using a mix of sample cDNAs as the template. It was determined from standard curves using the formula 10(-1/slope). For the calculations, the base of the exponential amplification function was used (e.g. 1.94 means 94% amplification efficiency). Relative expression levels were determined with efficiency correction, which considers differences in primer pair amplification efficiencies between target and reference gene [7-8]. Expression data and associated technical errors on duplicates were calculated using the gene expression module of the BIORAD iQ5 software, which follows models and error propagation rules outlined in the geNorm manual.
Statistical Analyzes
Relative expression levels were compared between yeast strains through analysis of variance (ANOVA) and significant differences were detected using the test of Tukey with a 0.05 threshold, using the STATISTICA software (99th Edition; Stat Soft).
Results and Discussion
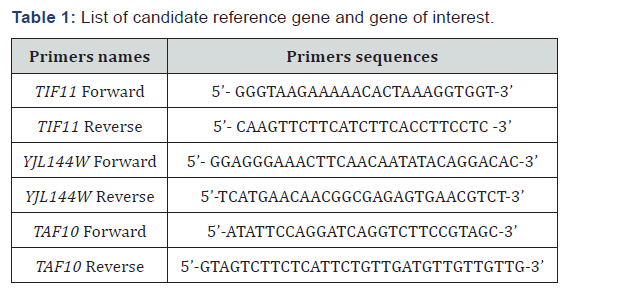
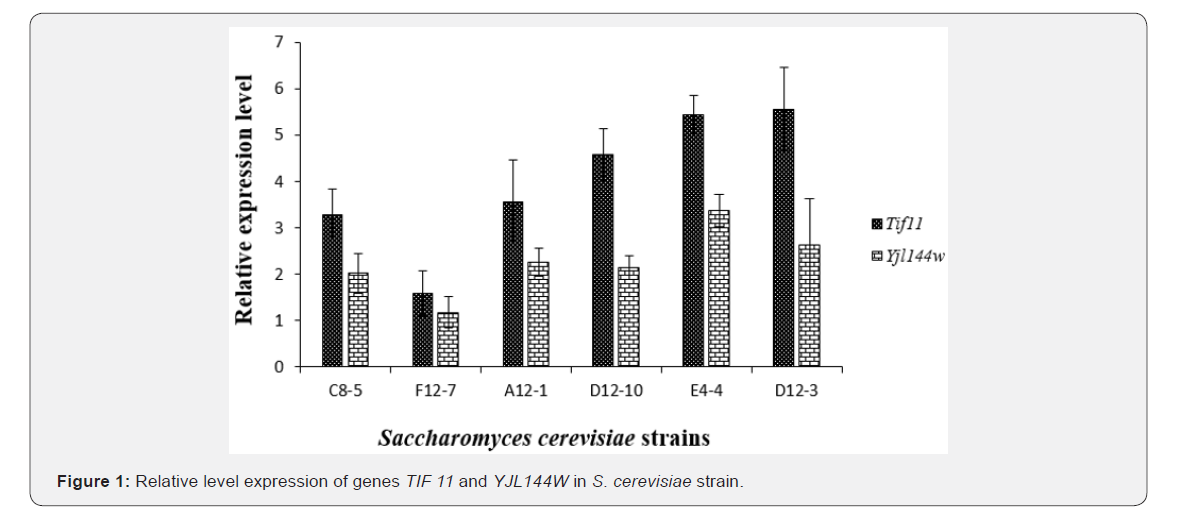
Cellular dehydration due to desiccation or drought is a common and potentially fatal stress encountered by many organisms including plants, animals and micro-organisms.Although adaptations to these constraints have been described physiologically and at the molecular level in many species, the functional significance of most adaptations was still uncertain. In this respect, the genes coding for hydrophilins, proteins linked to the stress due to the dehydration of the microorganisms, have been of particular interest. In recent years, they have been the subject of some studies to determine the levels of expression of these genes [1-9]. The ability of micro-organisms to adapt rapidly to changing environmental conditions is essential for their survival. Thus, in order to determine the suitability of yeast strains for the use in preparation of freeze-dried starters cultures, a study of genes involved in resistance to dehydration is necessary. In all the strains tested, the genes studied were expressed. Figures 1&2 showed the relative expression level of the TIF 11 and YJL144W genes respectively for Saccharomyces cerevisiae and Candida tropicalis strains compared to that of the reference gene of TAF 10. Generally, the TIF11 gene was more expressed than the YJL144W gene and this for all the strains of the two yeasts species studied. For Saccharomyces cerevisiae strains, the expression level of the TIF11 gene with respect to the highest internal TAF10 reference gene was obtained with strain D12-3 with an expression level of 5.56 ± 0.89 times greater.
Mean ± S.E.M = Mean values ± Standard error of means of three experiments.
Mean ± S.E.M = Mean values ± Standard error of means of three experiments.
The lowest expression was observed with strain F12-7 where the TIF11 gene was 1.59 ± 0.35 times expressed with respect to the reference internal gene TAF 10. For the YJL144W gene, the highest expression relative to the TAF 10 internal gene was observed with the E4-4 strain with a 3.37 ± 0.48-fold increase in expression relative to that of the TAF 10 gene. For this same gene, strain F12-7 was the one presenting the smallest expression 1.17 ± 0.33 times relative to the internal reference. For strains of Candida tropicalis, the TIF11 gene was 2.95 ± 0.12 times expressed in relation to the internal reference TAF 10 for the F0-5 strain. For the 2 others strains of the same species the expression was substantially similar with 2.59 ± 0.09 and 2.58±0.31 times that of TAF10 for C0-7 and C8-10 strains respectively. The YJL144W gene was more expressed in the C8-10 strain with an expression 2.23 ± 0.70 times greater than the TAF10 gene. For C0-7 and F0-5 strains, the expression levels of TIF11 and YJL144W are respectively 1.81 ± 0.06 and 1.78 ± 0.48 times higher than that of the TAF 10 gene. The statistical analysis revealed no difference between the strains for the 2 genes (P˃0.05).
For strains of Saccharomyces cerevisaie, the TIF11 gene is more expressed in the D12-3 strain, whereas for the YJL144W gene the expression is greater in the strain E4-4. In strain F12- 7, the TIF11 and YJL144W genes were the least expressed. The TIF11 and YJL144W genes have been reported to be important in strengthening the ability of yeasts to resist water stress [1- 9]. Our result was in line with those of Cordero-Otero et al. [9]. Indeed, these authors emphasized that among 12 genes tested only the genes TIF11 and RPL42, were overexpressed in the strain of Saccharomyces cerevisiae BY4742. Moreover, these genes are essential in the coding of the corresponding proteins TIF11p and RPL42p involved in maintaining cell viability. The work of Dang and Hincha, [1] showed that Saccharomyces cerevisiae cells in which the YJL144W and YMR175W genes were overexpressed were more tolerant to desiccation; which confirms the role of the two corresponding hydrophilins of the yeast in stress tolerance due to dehydration. Although the functional role of most hydrophilins has remained speculative, there was evidence to support their participation in acclimatization or adaptive response [10]. The ectopic expression of certain yeast hydrophilins confers tolerance to water deficiency conditions [11-12].
Also, as well as the genes have been expressed, the hydrophilins may not translated. However, the hydrophilin assay could give us more informations about their functions in yeasts. For Candida tropicalis strains, the statistical analyzes of the relative expression levels from the Tukey test revealed no difference between the strains for the 2 genes (P> 0.05). On basis of genes expression, the S. cerevisiae strains E 4-4 and D12-3 seemed appropriate to be used as freeze-dried cultures straters.
References
- Dang NX, Hincha DK (2011) Identification of two hydrophilins that contribute to the desiccation and freezing tolerance of yeast (Saccharomyces cerevisiae) cells. Cryobiology 62(3): 188-193.
- Hubalek Z (2003) Protectants used in the cryopreservation of microorganisms. Cryobiology 46(3): 205-229.
- Berny JF, Hennebert GL (1991) Viability and stability of yeast cells and filamentous fungus spores during freeze drying: effects of protectants and cooling rates. Mycologia 83(6): 805-815.
- Abadias M, Benabarre A, Teixidio N, Usall J, Vinas I (1001) Effect of freeze-drying and protectants on viability of the biocontrol yeast Candida sake. Int J Food Microbiol 65(3): 173-182.
- Zhao G, Zhang G (2005) Effect of protective agents, freezing temperature, and rehydration media on viability of malolactic bacteria subjected to freeze-drying. J Appl Microbiol 99(2): 333-338.
- N guessan KF, Brou K, Noemie J, Casaregola S, Djè KM (2011) Identification of yeasts during alcoholic fermentation of tchapalo, a traditional sorghum beer from Côte d’Ivoire. Antonie van Leeuwenhoek 99(4): 855-864.
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9): e45.
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4): 402-408.
- Cordero-Otero R, Gema LM, Boris RP, Margalef-Catala M (2012) The STF2p hydrophilin from Saccharomyces cerevisiae is required for dehydration stress tolerance. PLoS One 7(3): e33324.
- Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA, et al. (2008) The enigmatic LEA proteins and others hydrophilins. Plants Physiology 148(1): 6-24.
- Imai R, Chang L, Ohta A, Bray EA, Takagi M (1996) A lea-class gene of tomato confers salt and freezing tolerance when expressed in Saccharomyces cerevisiae. Gene 170(2): 243-248.
- Zhang L, Ohta A, Takagi M, Imai R (2000) Expression of plant group 2 and group 3 lea genes in Saccharomyces cerevisiae revealed functional divergence among LEA proteins. J Biochem 127(4): 611-616.






























