What is Host-Pathogen Initial Interaction Telling Us? an Essential Component of Biotic Stress Response Mechanism!
Pavitra Kotari1, Vinod Jatav2 and Ravishankar KundapuraV1*
1Department of Biotechnology, ICAR-Indian Institute of Horticultural Research, India
2Department of Vegetable Crops, ICAR-Indian Institute of Horticultural Research, India
Submission: December 01, 2018;Published: February 07, 2019
*Corresponding author: KV Ravishankar, Department of Biotechnology, ICAR-Indian Institute of Horticultural Research, Hesaraghatta Lake Post, Bengaluru 560 089, India
How to cite this article: Pavitra K, Vinod J, Ravishankar K. What is Host-Pathogen Initial Interaction Telling Us? an Essential Component of Biotic Stress Response Mechanism!. Adv Biotech & Micro. 2019; 12(4): 555843. DOI: 10.19080/AIBM.2019.12.555843
Abstract
Plants, in their natural environment are encountered by numerous pathogens. However, plants have evolved with the innate immune system that can deal with attack of these pathogens. Nevertheless, pathogens also keep modifying themselves to counter the host immunity and thus making the plants susceptible. Over the years, several studies in various crop species on plant pathogen interactions, showed that understanding of host-pathogens is still an intriguing area of investigation. In this review, we s discuss the recent studies, which show the active role of pathogens and their effector proteins in bringing up the disease development in the host system.
Keywords: Biotic stress; Plant pathogen; Signalling; Elicitors; Effectors; Disease development; Defense mechanism
Abbreviations: CWEDs: Cell Wall Degrading Enzymes; PAMPS: Pathogen Associated Molecular Patterns; PRRs: Pattern Recognition Receptors; PTI: Pattern Triggered Immunity; TALEs: Transcription Activator-Like Effectors; EBEs: Effector-Binding Elements; PR: Pathogenesis-Related
Introduction
Presently, biotic stress has become a global challenge, thereby significantly imposing intense selection pressure on plants leading to huge economic losses. Additionally, the changing climate is further worsening this situation. These stress factors (Bacteria, Fungi, Viruses, nematodes, insects and pests) act as un favourable conditions that bring about physiological and biochemical changes in plants resulting in inhibited growth, reduced economical yield, rendering acclimatization and adaptation of species to changing environment. Pathogens are those mediators that cause diseases in plants by manipulating the normal functioning of plant cells. There are mainly two types of pathogens Necrotrophs (Kill their host viz., Fusarium; Botrytis, Cochliobolus), and biotrophs (rarely kill their hosts viz., Erysiphe, Ustilago, Phytophthora) causing vascular wilts, leaf spots, cankers, wilting, stunting, chlorosis, malformations and affecting different parts of the plants.
Plant pathogens have evolved different strategies to infect their host and thus suppress host defense mechanism. By understanding the mode/mechanism of infection brought about by the pathogens would help to unravel the disease controlling strategies. Over the years lot of studies have been reported on plant pathogen interactions from both the pathogen and the plant viewpoint. Adapted pathogens circumvent and disturb host immunity, thus making plants susceptible. Sometimes, the failure of the host’s immune system also contributes towards plant disease susceptibility.
Host-pathogen interaction occurs on different levels during infection. Firstly, at the cell-wall level (First ground of battle field) and secondly at nucleus or cytoplasm level (Second ground of battle field). Pathogens, to make their entry into the host cell, make certain alterations in the plant cell walls by secreting different types of enzymes/elicitors that lead to destruction of cellulose, xylan and pectin eg. Cell Wall Degrading Enzymes (CWDEs). In turn, plants have evolved to combat the initial mode of infection by recognizing the Pathogen Associated Molecular Patterns (PAMPs) by Pattern Recognition Receptors (PRRs) leading to induce Pattern Triggered Immunity (PTI). In response to first line of defense, pathogens in turn modify them to bring about the infection at the second ground of battle field by secreting different types of virulence proteins through different secretion pathways.
Elicitors
Plants in their natural habitat exposed to several pathogens, releasing various chemical substances to exhibit a cascade of interactions between the host and its pathogen (Figure1). An elicitor is a molecule that triggers defense response in the plant system. They are diverse molecules eliciting the similar response in plants. Elicitors are basically of two types based on its origin, exogenous elicitors (pathogen origin) endogenous elicitors (plant origin). Elicitors are further divided into two categories based on its host interaction, General elicitors (infectious to all host) and Specific elicitors (infectious to specific host).
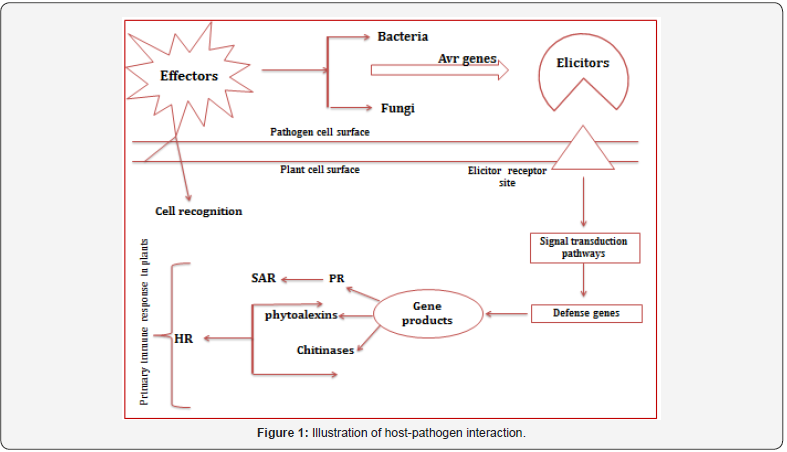
When pathogen encounters a host, it tries to overcome the important barrier (cell wall) that plants use to limit pathogen attack by producing enzymes that bring about the destruction of different components (Cellulose, xylan and pectin) of the cell wall resulting in establishment of pathogenesis. Recently, several studies have reported the role of elicitors upon pathogen infection. One such study reported describes about a novel protein CfPDIP1 secreted by Colletotrichum falcatum, exhibiting defense responses in sugarcane and triggering hypersensitive response in tobacco. Another study focused on β-glucans and chitin oligomers from Phytophthora and Pythium, Produced phytoalexins in soybean and rice plants and lignification in wheat leaves [1,2] studied the effect of oligogalacturonides from bacteria and fungi leading to synthesis of protein inhibitors and thus activating defense in Arabidopsis and tomato. Few studies have also been reported in viruses to elicit HR response in tobacco and tomato using viral coat protein hairpin from TMV [1]. Another report by [3], focused on Flagellin, flg 15 from Ralstonia bacteria, leading to deposition of callose and activation of defense genes in Arabidopsis.
Effectors
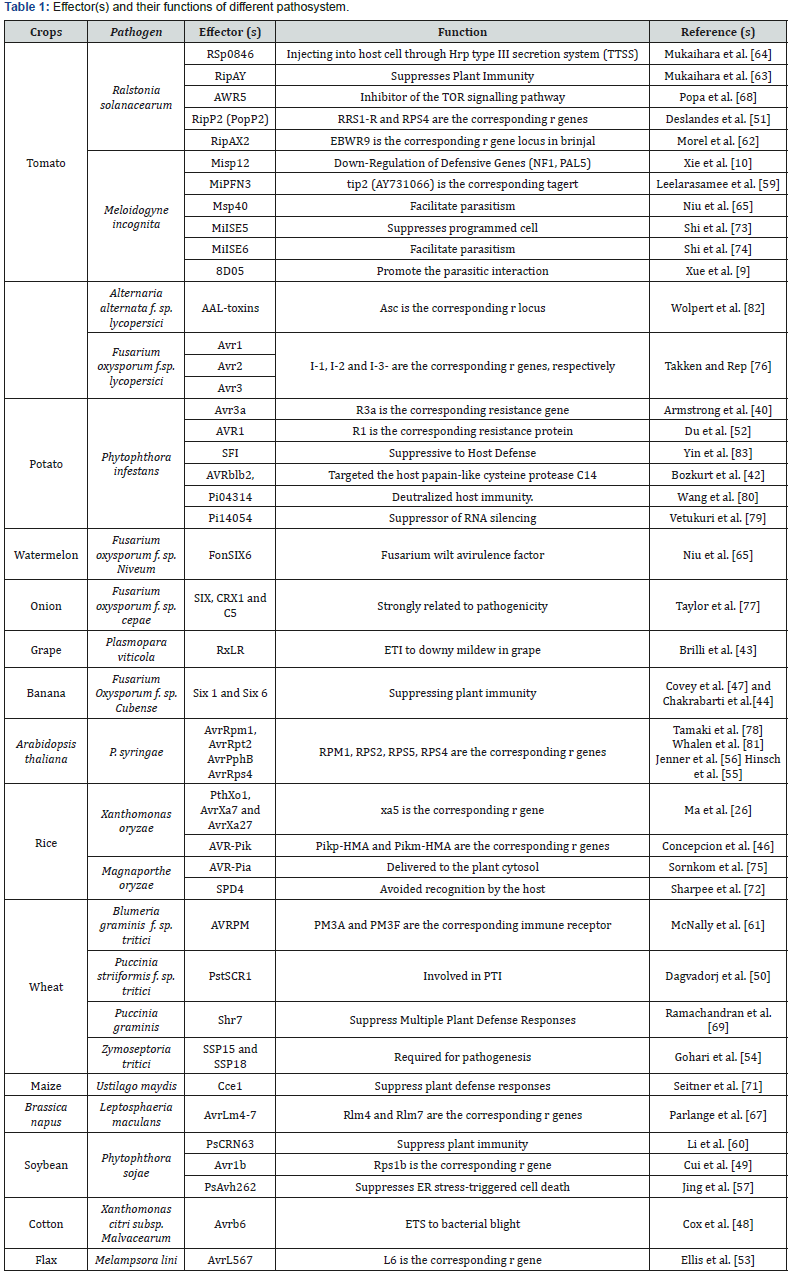
For the establishment of compatible interaction, pathogens must overcome Pattern Triggered Immunity (PTI) by secreting effector proteins that leads to Effector Triggered Susceptibility (ETS) and thereby be recognized by the host defense system, to exhibit Effector Triggered Immunity (ETI) [4]. In this review, we discuss the role of specific effectors of various pathogens affecting different crop species (Table 1).
Effectors are the protein molecules produced by virulence genes of pathogens. In order to initiate an infection, plant pathogens secrete a wide range of virulence proteins into the plant system through different types of secretion systems. There are different types of secretion pathways. Type I and II pathways secrete proteins to the host intercellular spaces, whereas type III and IV pathways deliver proteins or nucleic acids directly into the host cell. Examples of plant pathogen effectors secreted via the type I pathway are proteases and lipases from the softrot pathogen Erwinia chrysanthemi [5]. Type II secretion system is used by Ralstonia to translocate CWDEs. The prokaryotic pathogens (bacterium) mainly use a type three secretion system (T3SS).
While, the nematodes use the stylet to secrete effectors into the plant cells [6,7]. Further the fungal pathogens secrete the effectors into the plant apoplast or into the host cells and thus bring about the suppression of defense responses and alterations of the host metabolism. For example, the smut pathogen of maize, Ustilago maydis, encodes several hundreds of effector proteins that are virulence in nature and establish susceptibility [8]. Several nematode effectors responsible for parasitism have been reported by several researchers (Misp12, Msp40, 8D05) [9-13].
In order to counteract the effector molecules, plants have inculcated an inbuilt immune system to recognize effector proteins by producing specific R proteins (NLR receptors) [14]. A study reported by Gururania et al. [15] and Ali et al. [16] describe about the effector proteins from crop-infecting fungi, out of eightythree, 43 are encoded by Avr genes. Recent study, conducted by Bourras et al. [17] explains the effector protein (SvrPm3a1/f1) suppressing the immunity against powdery mildew pathogen in wheat.
Effectors have seen to perform diverse functions (pathogen self‐ defence, liberation of nutrients and manipulating signal transduction) as reported by Fatima and Senthil‐Kumar [18]. Example SnTox1 effector from Parastagonospora nodorum is seen to have multiple roles in host manipulation. Another Zt6 effector from the wheat pathogen Zymoseptoria tritici is seen to have phytotoxic and antimicrobial activity that affects both host and other microbes [19,20]. Another study reported by Rodriguez‐Moreno et al. [21] has described about the various infectious strategies used by the fungal pathogens to mediate host penetration, recognition escape and host defense inhibition.
Transcriptional Reprogramming
Recent transcriptomic studies demonstrated the fact that transcriptional changes in response to avirulent and virulent pathogens plays a vital role in both pathogenesis and resistance mechanism via up regulation or down regulation of the corresponding genes. Transcriptional changes in susceptible and resistant cultivars in response to pathogen, differ quantitatively bot not qualitatively [22]. Pathogen evolved to increase fitness (mutation in effector gene) in response of host by modulating host physiology in various ways. Some examples of diverse effector functions include modifying components of the immune system to evade detection and redirecting nutrients to the apoplast to support pathogen growth [23,24].
Transcriptional Reprogramming can broadly be classified into two groups. Firstly, effector proteins act as direct signal transducers, where the effector proteins are directly recognised by R- protein and thus induce the defense system of plant by up or down regulation of pathogenesis responsive genes. On the other hand, pathogens evolve to avoid recognition of effector protein by R-protein and hijack the transcriptional machinery of plants in favor of itself. For instance, Xanthomonas oryzae pv. oryzae injects Transcription Activator-Like Effectors (TALEs), directly into plant host cells, where they bind to specific promoter sequences in host genes (Xa5), which are designated as TAL Effector-Binding Elements (EBEs) [24,25], and promote the transcription of susceptibility (S) genes (OsSWEET11 and OsSWEET14). In response plant evolved naturally via mutating the Xa5 gene as xa5 (naturally occurring mutant form of Xa5) and these defective rice lines show reduced expression of the TALE-targeted susceptibility (S) genes, which results in increased resistance against bacterial blight in rice [26].
Secondly, effector proteins can act as indirect signal transducers; here the effector protein induces the signal transduction through other molecules. Virulent pathogens again evolved to use the plant transcription machinery according to their adaptation. For instance, CaCDPK15 was found to regulate pepper response to Pseudomonas solanacearum attack by indirectly activating CaWRKY40 expression, and CaWRKY40 in turn activated transcriptional expression of CaCDPK15 by binding to the CaCDPK15 [27]. Hyaloperonospora arabidopsidis, effector (HaRxL44), decreased SA immune processes by interfering with mediator function bringing enhanced susceptibility to downy mildew in arabidopsis [28]. Another previous report on Salicylic Acid (SA) required for the activation of genes encoding Pathogenesis-Related (PR) proteins such as PR1, potentiates the hypersensitive response triggered when pathogen effectors are directly or indirectly detected by R proteins [29]. Example. The biotrophic fungus Ustilago maydis causes smut disease in maize. Cmu1, an effector translocated into the cytoplasm of maize (Zea mays) plants by U. maydis, interferes with the SA pathway during disease development. Xanthomonas species have been shown to use a novel class of effectors, known as transcription activatorlike effectors, which function to bind specific host DNA sequences, resulting in the expression of host genes that aid pathogen colonization and infection [30,31]. Another review reported by Bezrutczyk et al. [32] has focused on the role of some pathogens that produce transcription activators, that induce the expression of sugar uptake transporters involved in bringing about plant susceptibility to pathogens.
Discussion and Conclusion
The co-evolution of plants and pathogens involves complex interaction mechanisms between the virulence factors (Elicitors and Effector proteins) of pathogens and the plants immune system (PTI and ETI). Cell wall being the initial battlefield for the pathogens and plants, both adopt contrasting wall-breaking and wall –reinforcing strategies to attack or to defend the entry of the foreign molecule [33].
Modification of cell walls is not the only strategy employed by fungal pathogens to prevent plant recognition. Where, pathogens make use of the CWDEs, as their first line of gadget of virulence to dissolve the cell wall components and establish the susceptible reaction/disease incidence. In response to this, plants also have developed a first line of defense mechanism known as basal immunity by recognizing the conserved microbial molecular patterns or PAMPs through PRRs leading to induction of PTI. An extensive review on plant Pattern Recognition Receptors (PRRs) role in detection of microbe‐associated molecular patterns and activation of intracellular signaling leading to PTI has been reported by Saijo et al. [34]. To overcome PTI induced defense, pathogens have further evolved with and developed different types of secretion systems to deliver and transport virulence factors (effectors) directly into the host system (cytosol) by bypassing / eluding the recognition facilitating the parasitism [35].
One such study reported, describes about the fungal pathogen that has evolved several strategies to overcome host immune responses by bringing about alterations in cell wall compositions and the secretion of effectors [36,37]. For instance, the tomato leaf mould fungus Cladosporium fulvum secretes the carbohydratebinding effector protein Ecp6 that suppresses chitin‐triggered host immunity [38].
In turn, to fight back, plants also have developed second line of defense (R gene mediated immunity and/or gene-gene recognition) by recognizing the effector proteins through specific, leading to induction of ETI. This is an ongoing process, in which pathogens keep evolving with new races and secreting modified effector proteins to increase their virulence. For example. Phytophthora infestans, encode RXLR‐type effectors that silence the activity of AVR3a that leads to a reduction in disease incidence [39-50].
Transcriptional re-programming is another gadget, that both pathogen and host make use of to exhibit a complex interaction mechanism. Here, both the pathogen and the host try to control transcription of different genes on a competition basis; the success decides the course of transcriptional mechanism for that particular gene. If the pathogen succeeds, the virulence gene(s)/gene product(s) bring about the re-programming in the expression of plant genes either by up-regulating susceptible genes or down-regulating resistant genes. On the other side, if the plant succeeds, it elicits the defense mechanism by bringing about the transcriptional re-programing of various defense related molecules (SA, JA and ET), R-genes and down regulation of susceptible genes. Several transcription factors (WRKY, ERF, AP2, bHLH, NAC, bZIP and MYB) also play a vital role in bringing about re-programing in the transcriptional mechanism [51-65].
Deciphering the role of pathogenicity factors (effector/ elicitor proteins) in the manipulation of host system would help us in understanding the mechanisms contributing towards the disease establishment and could also potentially lead to improve disease control methods. A thorough knowledge on the molecular mechanisms complementing with the virulence factors and receptor proteins would also lead to understand the gene for gene relationship among host and its pathogen, which finally would help in formulating pathogen specific resistance breeding programmes [66-83].
Acknowledgement
Authors thank the Indian Council of Agricultural Research, New Delhi for financial assistance through the ICAR Network Project on Transgenics in Crops: Functional Genomics-Fusarium wilt and drought tolerance in Banana.
Conflict of Interest
The authors declare no conflict of interest.
Informed consent
All authors give informed consent to publish the manuscript.
References
- Montesano M, Brader G, Palva ET (2003) Pathogen derived elicitors: searching for receptors in plants. Mol Plant Pathol 4(1): 73-79.
- Shibuya N, Minami E (2001) Oligosaccharide signalling for defence responses in plant. Physiological and Molecular Plant Pathology59(5): 223-233.
- Gómez‐Gómez L, Boller T (2000) An LRR‐ receptor‐like kinase involved in the perception of bacterial elicitor flagellin in Arabidopsis. Mol Cell 5(6): 1003-1011.
- Giraldo MC, Valent B (2013) Filamentous plant pathogen effectors in action. Nat Rev Microbiol 11(11): 800-814.
- Palacios JL, Zaror I, Martinez P, Uribe F, Opazo P, et al. (2001) Subset of hybrid eukaryotic proteins is exported by the type I secretion system of Erwinia chrysanthemi. Journal of Bacteriology 183(4): 1346-1358.
- Vieira P, Danchin EG, Neveu C, Crozat C, Jaubert S, et al. (2011) The plant apoplasm is an important recipient compartment for nematode secreted proteins. J Exp Bot 62(3): 1241-1253.
- Vanholme B, De Meutter J, Tytgat T, Van Montagu M, Coomans A, et al. (2004) Secretions of plant‐parasitic nematodes: a molecular update. Gene 332: 13-27.
- Lanver D, Tollot M, Schweizer G, Presti LL, Reissmann S, et al. (2017) Ustilago maydis effectors and their impact on virulence. Nat Rev Microbiol 15(7): 409-421.
- Xue B, Hamamouch N, Li C, Huang G, Hussey RS, et al. (2013) The 8D05 Parasitism Gene of Meloidogyne incognita Is Required for Successful Infection of Host Roots. Phytopathology 103(2): 175-181.
- Xie J, Li S, Mo C, Wang G, Xiao X, et al. (2016) A Novel Meloidogyne incognita Effector Misp12 Suppresses Plant Defense Response at Latter Stages of Nematode Parasitism. Front Plant Sci 7:964.
- Haegeman A, Mantelin S, Jones JT, Gheysen G (2012) Functional roles of effectors of plant-parasitic nematodes. Gene 492(1): 19-31.
- Hewezi T, Baum TJ (2013) Manipulation of plant cells by cyst and rootknot nematode effectors. Mol Plant Microbe Interact 26(1): 9-16.
- Mitchum MG, Hussey RS, Baum TJ, Wang X, Elling AA, et al. (2013) Nematode effector proteins: an emerging paradigm of parasitism. New Phytologist 199(4): 879-894.
- Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66: 487- 511.
- Gururania MA, Venkatesh J, Upadhyaya CP, Nookaraju A, Pandey SK, et al. (2012) Plant disease resistance genes: current status and future directions. Physiology and Molecular Plant Pathology 78: 51-65.
- Ali S, Laurie JD, Linning R, Cervantes-Chávez JA, Gaudet D, et al. (2014) An immunity-triggering effector from the Barley smut fungus Ustilago hordei resides in an Ustilaginaceae-specific cluster bearing signs of transposable element-assisted evolution. PLoS Pathog 10(7): e1004223.
- Bourras S, McNally KE, Müller MC, Wicker T, Keller B (2016) Avirulence genes in cereal powdery mildews: the gene-for-gene hypothesis 2.0. Front plant sci 7: 241.
- Fatima U, Senthil‐Kumar M (2015) Plant and pathogen nutrient acquisition strategies. Front Plant Sci 6: 750.
- Liu Z, Gao Y, Kim YM, Faris JD, Shelver WL, et al. (2016) SnTox1, a Parastagonospora nodorum necrotrophic effector, is a dual‐function protein that facilitates infection while protecting from wheat‐produced chitinases. New Phytol 211(3): 1052-1064.
- Kettles GJ, Bayon C, Sparks CA, Canning G, Kanyuka K, et al. (2017) Characterization of an antimicrobial and phytotoxic ribonuclease secreted by the fungal wheat pathogen Zymoseptoria tritici. New Phytol 217(1): 320-331.
- Rodriguez-Moreno L, Ebert MK, Bolton MD, Thomma BPHJ (2018) Tools of the crook‐infection strategies of fungal plant pathogens. Plant J 93(4): 664-674.
- Katagiri F, Tsuda K (2010) Understanding the plant immune system. Mol Plant Microbe Interact 23(12): 1531-1541.
- Block A, Alfano JR (2011) Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr Opin Microbiol 14(1): 39-46.
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, et al. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532.
- Mak AN, Bradley P, Bogdanove AJ, Stoddard BL (2013) TAL effectors: function, structure, engineering and applications. Curr opin in struct biol 23(1): 93-99.
- Ma W, Zou L, Zhiyuan JI, Xiameng XU, Zhengyin XU, et al. (2018) Xanthomonas oryzae pv. oryzae TALE proteins recruit OsTFIIAγ1 to compensate for the absence of OsTFIIAγ5 in bacterial blight in rice. Mol plant pathol 19(10): 2248-2262.
- Shen L, Yang S, Yang T, Liang J, Cheng W, et al. (2016) CaCDPK15 positively regulates pepper responses to Ralstonia solanacearum inoculation and forms a positive-feedback loop with CaWRKY40 to amplify defense signaling. Sci rep 6: 22439.
- Caillaud MC, Asai S, Rallapalli G, Piquerez S, Fabro G, et al. (2013) A downy mildew effector attenuates salicylic acid–triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS biology 11(12): e1001732.
- Brodersen P, Malinovsky FG, Hématy K, Newman M A, Mundy J (2005) The role of salicylic acid in the induction of cell death in Arabidopsis acd11. Plant Physiol 138(2):1037-1045.
- Boch J, Scholze H, Schornack, S, Landgraf A, Hahn S, et al. (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326(5959): 1509-1512.
- Moscou MJ, Bogdanove A J (2009) A simple cipher governs DNA recognition by TAL effectors. Science 326(5959): 1501-1501.
- Bezrutczyk M, Yang J, Eom JS, Prior M, Sosso D, et al. (2018) Sugar flux and signaling in plant-microbe interactions. Plant J 93(4): 675-685.
- Ryder LS, Talbot NJ (2015) Regulation of appressorium development in pathogenic fungi. Curr Opin Plant Biol 26: 8-13.
- Saijo Y, Loo E PI, Yasuda S (2018) Pattern recognition receptors and signaling in plant-microbe interactions. Plant J 93(4): 592-613.
- Salmond GP (1994) Secretion of extracellular virulence factors by plant pathogenic bacteria. Annual review of phytopathology 32(1): 181-200.
- Rovenich H, Boshoven JC, Thomma BPHJ (2014) Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Curr Opin Plant Biol 20: 96-103.
- Sánchez-Vallet A, Mesters JR, Thomma BP (2015) The battle for chitin recognition in plant-microbe interactions. FEMS microbiol Rev 39(2): 171-183.
- Sánchez-Vallet A, Saleem-Batcha R, Kombrink A, Hansen G, Valkenburg DJ, et al. (2013) Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. Elife 2: e00790.
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, et al. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450(7166): 115-118.
- Armstrong MR, Whisson SC, Pritchard L, Bos JI, Venter E, et al. (2005) An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci 102(21): 7766-7771.
- Ashwin NMR, Barnabas L, Ramesh Sundar A, Malathi P, Viswanathan R, et al. (2018) CfPDIP1, a novel secreted protein of Colletotrichum falcatum, elicits defense responses in sugarcane and triggers hypersensitive response in tobacco. Appl Microbiol Biotechnol 102(14): 6001-6021.
- Bozkurt TO, Schornack S, Win J, Shindo T, Ilyas M, et al. (2011) Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. PNAS 108 (51): 20832-20837.
- Brilli M, Asquini E, Moser M, Bianchedi PL, Perazzolli M, et al. (2018) A multi-omics study of the grapevine-downy mildew (Plasmopara viticola) pathosystem unveils a complex protein coding and noncoding- based arms race during infection. Scientific Reports 8:757.
- Chakrabarti A, Ganapathi TR, Mukherjee P K, Bapat VA (2003) MSI-99, a magainin analogue, imparts enhanced disease resistance in transgenic tobacco and banana. Planta 216(4): 587-596.
- Cohn J, Sessa G, Martin GB (2001) Innate immunity in plants. Curr Opin Immunol 13(1): 55-62.
- Concepcion JC, Franceschetti M, Maqbool A, Saitoh H, Terauchi R, et al. (2018) Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen. Nature Plants 4: 576-585.
- Covey PA, Kuwitzky B, Hanson M, Webb KM (2014) Multilocus analysis using putative fungal effectors to describe a population of Fusarium oxysporum from sugar beet. Phytopathology 104(8): 886-896.
- Cox KL, Meng F, Wilkins KE, Li F, Wang P, et al. (2017) TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton. Nat Commun 8: 15588.
- Cui L, Yin W, Dong S, Wang Y (2012) Analysis of polymorphism and transcription of the effector gene Avr1b in Phytophthora sojae isolates from China virulent to Rps1b. Mol Plant Pathol 13(2):114-122.
- Dagvadorj B, Ozketen AC, Andac A, Duggan C, Osman T, et al. (2017) A Puccinia striiformis f. sp. tritici secreted protein activates plant immunity at the cell surface. Scientific Reports, 7: 1141.
- Deslandes L, Olivier J, Theulieres F, Hirsch J, Feng DX, et al. (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci U S A 99(4): 2404-2409.
- Du Y, Weide R, Zhao Z, Msimuko P, Govers F, et al. (2018) RXLR effector diversity in Phytophthora infestans isolates determines recognition by potato resistance proteins; the case study AVR1 and R1. Studies in mycology 89: 85-93.
- Ellis JG, Dodds PN, Lawrence GJ (2007) Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu Rev Phytopathol 45: 289-306.
- Mirzadi Gohari A, Ware SB, Wittenberg AH, Mehrabi R, Ben M’Barek S, et al. (2015) Effector discovery in the fungal wheat pathogen Zymoseptoria tritici. Mol plant pathol 16(9): 931-945.
- Hinsch M, Staskawicz BJ (1996) Identification of a new Arabidopsis disease resistance locus, RPS4, and cloning of the corresponding avirulence gene, AvrRps4, from Pseudomonas syringae pv. pisi. Mol Plant Microbe Int 9(1): 55-61.
- Jenner C, Hitchin E, Mansfield J, Walters K, Betteridge P, et al. (1991) Gene-for gene interactions between Pseudomonas syringae pv. phaseolicola and Phaseolus. Mol Plant Microbe Interact 4(6): 553-562.
- Jing M, Guo B, Li H, Yang B, Wang H, et al. (2016) A Phytophthora sojae effector suppresses endoplasmic reticulum stress-mediated immunity by stabilizing plant binding immunoglobulin proteins. Nat commun 7:11685.
- King R, Urban M, Hammond-Kosack MC, Hassani-Pak K, Hammond-Kosack KE (2015) The completed genome sequence of the pathogenic ascomycete fungus Fusarium graminearum BMC Genom 16: 544.
- Leelarasamee N, Zhang L, Gleason C (2018) The root-knot nematode effector MiPFN3 disrupts plant actin filaments and promotes parasitism. PLoS Pathog, 14(3): 1006947.
- Li Q, Zhang M, Shen D, Liu T, Chen Y, et al. (2016) A Phytophthora sojae effector PsCRN63 forms homo-/heterodimers to suppress plant immunity via an inverted association manner. Sci Rep 6:26951.
- McNally KE, Menardo F, Lüthi L, Praz CR, Müller MC (2018) Distinct domains of the AVRPM3A2/F2 avirulence protein from wheat powdery mildew are involved in immune receptor recognition and putative effector function. New Phytol 218(2): 681-695.
- Morel A, Guinard J, Lonjon F, Sujeeun L, Barberis P, et al. (2018) The eggplant AG91-25 recognizes the Type III-secreted effector RipAX2 to trigger resistance to bacterial wilt (Ralstonia solanacearum species complex). Mol Plant Pathol 19(11): 2459-2472.
- Mukaihara T, Hatanaka T, Nakano M and Oda K (2016) Ralstonia solanacearum type III effector RipAY is a glutathione-degrading enzyme that is activated by plant cytosolic thioredoxins and suppresses plant immunity. M Bio 7(2): 359-416.
- Mukaihara T, Tamura N (2009) Identification of novel Ralstonia solanacearum type III effector proteins through translocation analysis of hrpB-regulated gene products. Microbiology 155(7): 2235-2244.
- Niu J, Liu P, Liu Q, Chen C, Guo Q, et al. (2016) Msp40 effector of rootknot nematode manipulates plant immunity to facilitate parasitism. Scientific Reports 6: 19443.
- Niu X, Zhao X, Ling KS, Levi A, Sun Y, et al. (2016) The FonSIX6 gene acts as an avirulence effector in the Fusarium oxysporum f. sp. niveum - watermelon pathosystem. Sci Rep 6: 28146.
- Parlange F, Daverdin G, Fudal I, Kuhn ML, Balesdent MH, et al. (2009) Leptosphaeria maculans avirulence gene AvrLm4-7 confers a dual recognition specificity by the Rlm4 and Rlm7 resistance genes of oilseed rape, and circumvents Rlm4-mediated recognition through a single amino acid change. Mol Microbiol 71(4): 851-863.
- Popa C, Li L, Gil S, Tatjer L, Hashii K, et al. (2016) The effector AWR5 from the plant pathogen Ralstonia solanacearum is an inhibitor of the TOR signalling pathway. Scientific reports 6: 27058.
- Ramachandran SR, Yin C, Kud J, Tanaka K, Mahoney AK, et al. (2017) Effectors from wheat rust fungi suppress multiple plant defense responses. Phytopathology 107(1): 75-83.
- Raffaele S, Farrer RA, Cano LM, Studholme DJ, MacLean D, et al. (2010) Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330(6010): 1540-1543.
- Seitner D, Uhse S, Gallei M, Djamei A (2018) The core effector Cce1 is required for early infection of maize by Ustilago maydis. Mol Plant Pathol 19(10): 2277-2287.
- Sharpee W, Oh Y, Yi M, Franck W, Eyre A, et al. (2017) Identification and characterization of suppressors of plant cell death (SPD) effectors from Magnaporthe oryzae. Mol plant pathol 18(6): 850-863.
- Shi Q, Mao Z, Zhang X, Ling J, Lin R, et al. (2018) The Novel Secreted Meloidogyne incognita Effector MiISE6 Targets the Host Nucleus and Facilitates Parasitism in Arabidopsis. Front Plant Sci 9: 252.
- Shi Q, Mao Z, Zhang X, Zhang X, Wang Y, et al. (2018) A Meloidogyne incognita effector MiISE5 suppresses programmed cell death to promote parasitism in host plant. Scientific Reports 8: 7256.
- Sornkom W, Miki S, Takeuchi S, Abe A, Asano K, et al. (2017) Fluorescent reporter analysis revealed the timing and localization of AVR-Pia expression, an avirulence effector of Magnaporthe oryzae. Mol Plant Pathol 18(8): 1138-1149.
- Takken F, Rep M (2010) The arms race between tomato and Fusarium oxysporum. Mol plant pathology 11(2): 309-314.
- Taylor A, Vágány V, Jackson AC, Harrison RJ, Rainoni A, et al. (2016) Identification of pathogenicity‐related genes in Fusarium oxysporum f. sp. cepae. Mol plant pathol 17(7): 1032-1047.
- Tamaki S, Dahlbeck D, Staskawicz B, Keen NT (1988) Characterization and expression of two avirulence genes cloned from Pseudomonas syringae pv. Glycinea. J Bacteriol 170(10): 4846-54.
- Vetukuri RR, Whisson SC, Grenville-Briggs LJ (2017) Phytophthora infestans effector Pi14054 is a novel candidate suppressor of host silencing mechanisms. European Journal of Plant Pathology 149(3): 771-777.
- Wang S, Boevink PC, Welsh L, Zhang R, Whisson SC, et al. (2017) Delivery of cytoplasmic and apoplastic effectors from Phytophthora infestans haustoria by distinct secretion pathways. New Phytol 216(1): 205-215.
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ (1991) Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3(1): 49-59.
- Wolpert TJ, Dunkle LD, Ciuffetti LM (2002) Host-selective toxins and avirulence determinants: What’s in a name? Annu Rev Phytopathol 40: 251-285.
- Yin J, Gu B, Huang G, Tian Y, Quan J, et al. (2017) Conserved RXLR Effector Genes of Phytophthora infestans Expressed at the Early Stage of Potato Infection Are Suppressive to Host Defense. Front Plant Sci 8: 2155.






























