An in-Silico Approach to Understanding the Structure-Function: A Molecular Dynamics Simulation Study of Rand Serine Protease Properties from Bacillus Subtilis in Aqueous Solvents
Randa Abdel Kareem Abusham1,2, Malihe Masomian1,2, Abu Bakar Salleh1, Adam Thean Chor Leow1,3 and Raja Noor Zaliha Raja Abd Rahman1,2 *
1Enzyme and Microbial Technology Research Center, University Putra Malaysia, Malaysia
2Department of Microbiology, University Putra Malaysia, Malaysia
3Department of Cell and Molecular Biology, University Putra Malaysia, Malaysia
Submission: November 12, 2018; Published: January 03, 2019
*Corresponding author: Raja Noor Zaliha Raja Abd Rahman, Enzyme and Microbial Technology Research Center, Faculty of Biotechnology and Biomolecular Sciences, University Putra Malaysia, 43400 UPM, Serdang, Selangor, Malaysia
How to cite this article: Randa AKA, Malihe M, Abu BS, Adam TCL, Raja NZRAR. An in-Silico Approach to Understanding the Structure-Function: A Molecular Dynamics Simulation Study of Rand Serine Protease Properties from Bacillus Subtilis in Aqueous Solvents. Adv Biotech & Micro. 2019; 12(1): 555828. DOI: 10.19080/AIBM.2019.12.555828
Abstract
The protein’s 3D structure is important for designing a novel biocatalyst that suits industrial applications. Moreover, the data from the 3D structure may increase enzyme production and help to analyse and provide an understanding of the structure of the thermostable, organic-solvent-tolerant Rand serine protease. A structural prediction for the Rand serine protease was built using the Yet Another Scientific Artificial Reality Application (YASARA) structure employing a known 3D structure (subtilisin E-propeptide complex; PDB ID: 1SCJ) as a template, which has the highest sequence identity (96%) to the Rand serine protease. In silico study of docking, the substrate N-succinyl-alanyl-alanyl-prolyl-phenylalanine-4-nitroanilide in Rand protease resulted in 25 clusters whereby 4 clusters observed to involve the catalytic triad of rand protease -. Molecular dynamics (MD) simulations were performed in two organic solvents with different logP values, such as pyridine (logP 0.71), benzene (logP 2.0) and pure water, for 10 ns to investigate their effect on the Rand protease structure. The predicted 3D structure of the Rand protease revealed that the topological organization of the α/β-hydrolase fold consisted of 6 α-helices and 13 β-strands. The active site of the enzyme predicted to be consisted of three amino acids residues Asp32, His64 and Ser221. This docking result was in good agreement with the active site prediction - study, which showed the same conserved catalytic triad. In conclusion, the structure analysis confirmed the unique characteristics of this enzyme and explained the organic solvent stability of the enzyme.
Keywords: Rand protease; serine proteases; structure; active sites; docking; molecular dynamics
Abbreviations: YASARA: Yet Another Scientific Artificial Reality Application; NCBI: National Center for Biotechnology Information; MTDC: Malaysian Technology Development Centre
Introduction
Serine proteases from the Bacillus species extensively applied in the biotechnological application [1,2] such as detergent, leather and food industries, frequently under non-physiological conditions. New proteases with improved performance at extreme temperatures and in the presence of chemical additives may have great economic potential. The increasing availability of genetic sequences from completely different environments makes homology-based screening an attractive strategy for the discovery of new proteases [3]. So far, the broad investigation on proteases gave the basic understanding of their catalytic mechanism and their structure-function. Computational structure analysis and homology modeling can be a key process for the 3D structure reconstruction which facilitates the protein-protein interaction research. Protein crystal is the basic necessity to obtain the 3D structure [4].
A computational simulation is an important tool that is used to solve scientific problems by performing numerical experiments on computers. It elucidates how a complex system works without synthesising it. It can capture the system’s details, which may be invisible in a bench experiment because of the limitations of the technique. In addition, it can be used as a predictive tool that aids the design of the experiment and also helps to save on the cost. MD is among the popular approaches for computational simulation [5]. Generally, the principles governing the activation and inhibition effects of organic solvents towards enzymes have long remained ambiguous; however, due to the reported significant effort put into understanding enzyme-solvent interactions, some probable reasons and therefore the underlying causes can now be understood [6].
In order to provide a better understanding of the protein function, structure prediction by employing homology modeling was conducted. Protein crystal is the basic necessity of obtaining the 3D structure. Alternatively, the best strategy to get that information is to predict the enzyme’s 3D structure. In the case of enzymes, determination of 3D structures has helped to elucidate why residues far apart in the sequence are involved in a given catalytic reaction [7].In addition, the close relationship between the protein’s structure and function is manifested in the lock-andkey hypothesis and induced-fit model of protein binding, where the protein structure is fixed or locally adjusted when a ligand binds to the active site [8].
Serine proteases comprise two discrete families: bacterial serine proteases (e.g. subtilisin) and mammalian serine proteases (e.g. chymotrypsin, trypsin and elastase). They differ in terms of amino acids sequences and 3D structure, despite having a common active site geometry and enzymatic mechanism [9]. Subtilisin’s are a family of alkaline serine end proteases secreted by a wide variety of Bacillus species: subtilisin E from Bacillus Subtilis 1168 [10], Tk-subtilisin from Thermococcus kodakaraensis [11], a subtilisin-like serine protease from Bacillus Subtilis DM-04 [12], subtilisin E from B. subtilis, and subtilisin nattokinase from Bacillus Subtilis var. natto [13]. The enzyme having no disulphide bonds consists of a single polypeptide chain of about 275 amino acid residues (molecular was about 27,500 Da), and possesses the ‘catalytic triad’ of Asp, His and Ser residues, which is conserved among serine proteases, as the hallmark of its active sites [14].
Rand protease is classified as serine protease and it is a thermostable and organic solvent tolerant enzyme with broad applications. However, a study on the structure of such an enzyme is limited. Three-dimensional (3D) structure of a protein is needed to understand their properties. The structural stability of the enzyme is an important determinant of the catalytic activity in organic solvents [15]. Therefore, this project studied the effect of different organic solvents on the structure and flexibility of the Rand protease.
Materials and Methods
The amino acid composition analysis
The amino acid sequence of Rand protease was analysed by ExPASy – Prot Param tool.
Structure prediction
Model building: The prediction of the Rand protease structure was conducted using automated homology modelling by YASARA (10.4.11) [16]. The template search was performed by aligning the target sequence with other bacterial proteases using PSI-BLAST from NCBI based on the PDB database. The top homologues were ranked based on the alignment score and structure quality. Subtilisin-propeptide complex (PDB ID: 1SCJ) with a sequence identity of 96% was selected and used as the template. Backbone, loop and side chains were then built, optimised and fine-tuned.
Model refinement: An unrestrained, high-resolution refinement with explicit solvent molecules was run, using YASARA 03 force fields. The result was validated by the YASARA program to ensure that the refinement did not move the model in the wrong direction, based on the template used to predict the structure. For each template, five models were built. To refine the geometry and orientation of the binding model, the highest-ranked models for each template were subsequently refined using the YASARA program. In the refinement, after the side chains had been built, optimised and fine-tuned, all the newly modelled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of the aligned residues were kept fixed to avoid potential damage) until the force field energy converged. Then, YASARA combined the best parts of the models to obtain a hybrid model, to increase the accuracy beyond that of each of the contributors. Finally, the model was subjected to a final round of simulated annealing minimization in an explicit solvent and the quality Z-scores were obtained fully automatically.
Model quality and accuracy
The stereochemical quality of the model-built structure, the accuracy of parameters such as bond lengths, bond angles and torsion angles, and the correctness of the amino acid chirality were evaluated using the Precheck and Errata programs. The PDB file for the predicted structure was submitted in the aforementioned website, and the results were analysed and discussed.
Active site prediction
The active site prediction in the refined model was done by submitting the Rand protease amino acid sequences to.
In silico study
Molecular docking: Predictions of Rand protease towards the substrate were performed using in silico experimental approach. The molecular docking task was conducted using the Yet Another Scientific Artificial Reality Application (YASARA) Structure package [16]. Docking was also conducted to determine the binding site of the substrate. The substrate used was N-succinylalanyl- alanyl-prolyl-phenylalanine-4-nitroanilide and the structure was obtained from PubChem under the National Center for Biotechnology Information (NCBI) PubChem CID 5496888.
Molecular Dynamics (MD) simulations: MD simulation techniques were used to analyse the dynamic behavior of the Rand protease, and different organic solvents with different log P (pyridine (log P 0.71) and benzene (log P 2.0)); water was used as the control. The complex of the Rand protease and the organic solvents were used for MD simulation. MD simulation was conducted using the YASARA program [17]. The simulation was carried out in an explicit water environment, at a constant pressure, using an AMBER03 force field, in a periodic cell boundary condition and the model was simulated at 295 K (25 °C). The solvated structure was minimized by the steepest descent method for 15,000 steps at a temperature of 295K and a constant pressure. Then the complex was equilibrated for a 2ns period. After equilibration, a production MD was run for 10 ns at a constant temperature and pressure. The RMSD and RMSF values were analysed for each simulation conducted. The RMSF for the average equilibrium conformation is often used as an indicator of protein rigidity, which is considered to be a feature that is related to the resistance to unfolding [18]. All the prediction and simulation studies were done on a Dell PC and a rack/blade server.
Results and Discussion
The amino acid composition of the Rand protease full length
The ORF Rand protease protein consists of 381 amino acids. Analysis of the amino acid sequence of the Rand protease reveals that this protein is rich in hydrophobic amino acids (49.4%). Ala, Gly, Val, Leu, Ile, Pro, Phe, Met and Trp were present in high concentrations at percentages of 12.6%, 10.0%, 9.2, 5.5%, 5.0%, 3.7%, 2.4% and 1.0%, respectively. The amount of negatively charged residues (Asp + Glu) is 7.7% and the amount of positively charged residues (Arg + Lys) is 6.8%. Hence, the total charged residues are 14.5% of the whole protein (Table 1). No cysteine residues were found in the sequence, indicating that the protein cannot form a disulfide bridge. The highest numbers of amino acid residues are of Ala, Ser, Gly, Thr and Val, which account for 47.1% of total residues.
Their common feature is their smaller size. Serine (Ser), one of the highest in number, is known to confer thermostability by forming additional intramolecular hydrogen bonds, such as those observed in the critical places of triosephosphate isomerase [19]. Different amino acids play a different role in determining a protein’s structure and function. In this case, the factors responsible for protein stability based on differences in amino acids was studied by Pack & Yoo [20]. The higher alanine content in thermophilic proteins is believed to reflect the fact that Ala is the best helix-forming residue [21].
Structure prediction
Template search and selection: Structure prediction by homology modelling is based on accurate sequence alignments between a query protein and a template protein with solved structures; hence, the prediction accuracy heavily depends on the sequence similarity between the two proteins [22]. The prediction of the Rand protease structure has been done on the basis of the respective amino acid sequences. Rand protease sequence was found to be 96% identical to subtilisin E, for which the 3D structure is available [23]. Homology models are built on the basis of a significant sequence identity between target and template, as an alignment with more than 50% sequence identity generally contains almost no error [22,24]. For closely related protein sequences with identities higher than 30%–40%, the alignments produced by all methods are always correct [25].
The critical first step in structure prediction is the identification of the best template structure. Nowadays, there are sensitive methods based on multiple sequence alignment that could help to find distant evolutionary relationships; one of which, PSI-BLAST, constructs and performs an NCBI BLAST search with a custom, position-specific, scoring matrix. The amino acid sequence of the Rand protease with a protein ID of AEG88964 was aligned using PSI-BLAST at the NCBI. BLAST was developed to perform rapid searches for homologous sequences in PDB. The results from PSI-BLAST show that the Rand protease exhibits a high similarity to the crystal structures of the subtilisin propeptide from Bacillus Subtilis (1SCJ_A), nattokinase from Bacillus Subtilis natto (4DWW_A), pro-subtilisin E from Bacillus Subtilis subsp (3WH1_A) and subtilisin from Bacillus pumilus (1MEE_A).
Model building: The Rand serine protease contains 275 amino acid residues consisting of nine beta sheets and nine alpha helices, as shown in Figure 1A. The topology of a protein structure is a highly simplified description of its fold, including only the sequence of secondary structure elements, and their relative spatial positions and approximate orientations. These topology cartoons are useful for understanding particular folds and making comparisons between similar subtilisin’s’ folds. From this topology diagram, it is observed that the StmPr1 protease has the α /β fold catalytic center containing the typical seven-stranded parallel β-sheet, which is observed with other subtilisin’s [26].
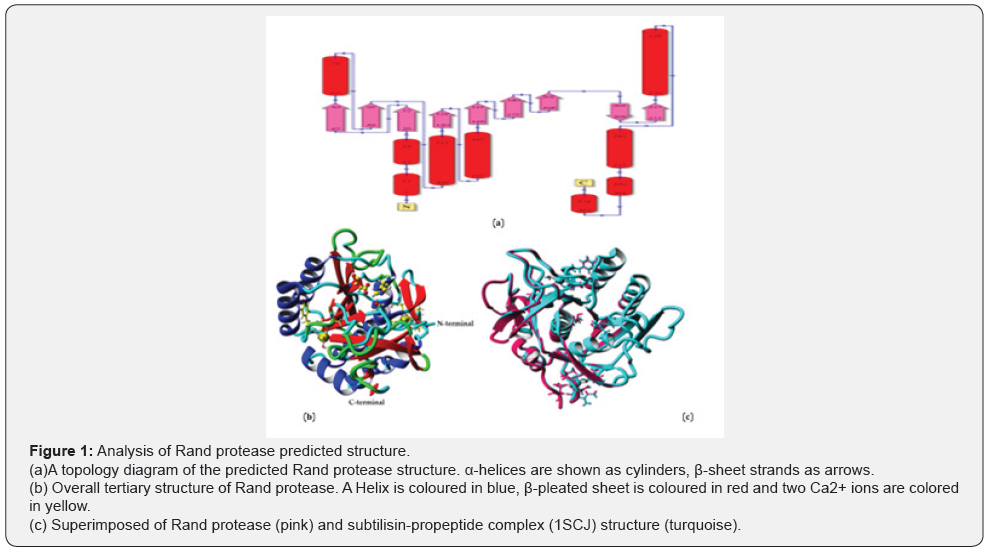
The analysis of the secondary structure indicates that the Rand protease accommodates 33.1% of α-helix, 22.5% of ß-sheet, 17.1% turn, 27.3% coil and 0.0% 3-10 helix. The overall tertiary structure of the Rand protease is shown in Figure 1B. The superimposed image of the Rand protease structure with the subtilisin-propeptide complex (1SCJ) structure demonstrates an RMSD value of 0.036 Å over 3,849 matched atoms (Figure 1C), which indicate the slight difference between the atoms in the structures of the two proteases. In general, an RMSD value that is less than 3Å implies a strong similarity between the structures [27] (Figure1).
Model validation: The quality evaluation of the predicted model structure could determine the information on the accuracy of 3D protein models. To verify the stereochemical the quality evaluation of the predicted model structure could determine the information about the accuracy of the 3D protein models. To verify the stereochemical quality of the model-built structure, the accuracy of parameters such as bond lengths, bond angles, torsion angles and the correctness of the amino acid chirality have to be evaluated. Many alternative programs and servers are available for model evaluation, some of which are listed in (Table 2).
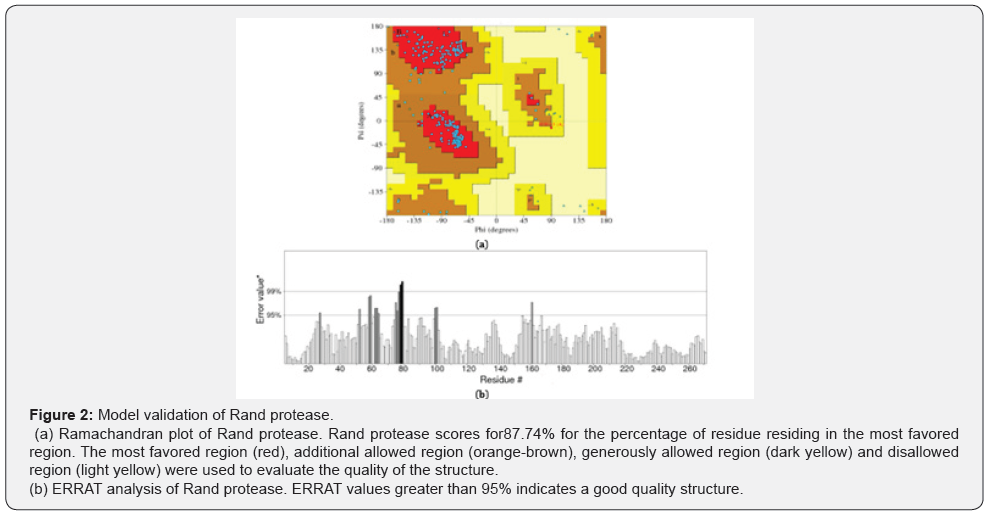
The quality and accuracy of the Rand protease structure were evaluated using a Ramachandran plot (Figure 2A). Ramachandran plot, also known as Ramachandran diagram, is an implement for the analysis of a protein structure. It calculates the phi and psi values for each residue in a protein [28]. The white areas in the Ramachandran diagram are sterically disallowed for all amino acids besides glycine.
The Ramachandran plot illustrates that 87.7% of the residues lie in the most-favored region, which is close to a good structure, while 11.9% are in the additional favored region and 0.4% are in generously allowed region. However, there are no residues found in the disallowed region (Figure 2). ERRAT is a protein structure tool used to evaluate the progress of crystallographic model building and refinement. The ERRAT program evaluates the statistics of non-bonded atom-atom interaction such as the distance between carbon-carbon, carbon-nitrogen, carbonoxygen, nitrogen-nitrogen, nitrogen-oxygen and oxygen-oxygen bonds [29]. The ERRAT analysis shows that the overall quality of the Rand protease was 94.4% (Figure 2B), which is near to the good quality structure. The quality analysis of protein structures is a crucial element in structure validation study and plays an important role in protein structure prediction, where the predicted model may contain significant errors [30] and it shows the reliability of the structure for further study.
Active site prediction
The prediction of active sites using the prediction model was done by submitting the Rand protease amino acids sequence to (Figure 3A). The 3D structure is complex and compact, which is suited to the thermostability properties of Rand protease. The compactness of the 3D structure is higher in comparison to the psychrophilic enzyme. The catalytic triad proposed for Rand protease was His64, Asp32 and Ser221 (Figure 3B) which is suggested to be responsible for catalytic properties of Rand protease.
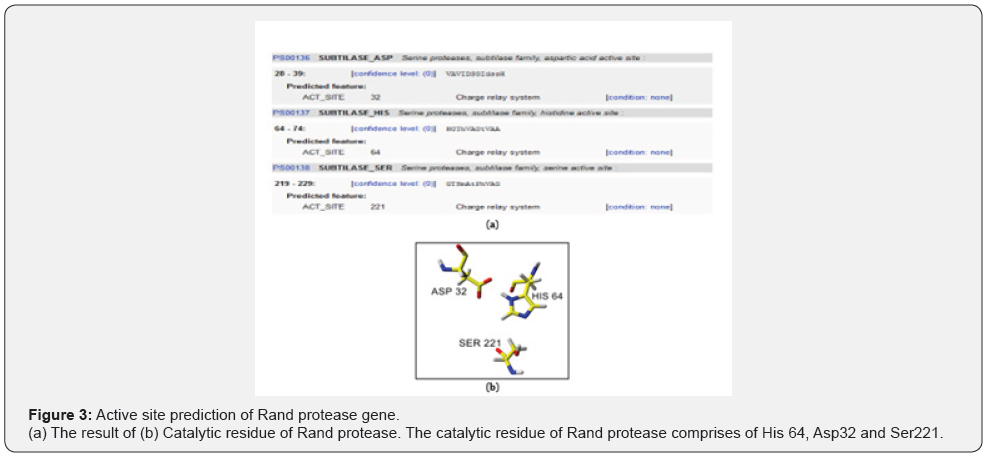
The coordinates of subtilisin BPN’, subtilisin Carlsberg, thermitase, Savinase, Esperase and Proteinase K were used previously by Siezen et al. [31]. This core of about 190 residues contains virtually all of the common α-helix and β-strand elements, including the active site residues D-32, H-64 and S-221.
Serine proteases are characterized by the catalytic triad of aspartic acid, histidine and serine residues [32]. The catalytic triads for a subtilisin-like serine protease produced by thermophilic bacterium Fervid bacterium pennivorans are Asp41, His79 and Ser260. The numbering order begins at the start of the mature protease [33]. SapSh exhibits sequence similarities with members of the subtilisin family of proteases, and there is a high level of conservation in the regions around a putative catalytic triad consisting of Asp30, His65 and Ser369[34]. The subtilisinlike proteases exhibit a catalytic triad consisting of Asp30, His94 and Ser415 [35].
A variation of the Ser/His/Asp triad is found in the aspartyl dipeptidase protease, where the aspartate residue is replaced by a glutamate residue [36]. While aspartyl dipeptidase produced by salmonella typhimurium belongs to a new serine protease family with a Ser/His/Glu triad [37]. Another different site triad, Ser/ His/His, is found in herpes virus proteases that play a key role in the maturation of the assembly protein. The cytomegalovirus protease is inactivated by DFP. Ser132 was known as the active site serine of the human cytomegalovirus protease because it was modified by the inhibitor. The invariant His63 is critical for activity as determined by site-directed mutagenesis [38].

Calcium binding domain: Calcium binding sites in proteins are known to stabilize the active form of the protein, and also assist in the activation of the catalytic triad of protein and regulatory processes. There are two calcium binding sites found in the Rand protease structure. The coordination of the Ca2+ 277 composed of Ala169, Tyr171, Thr174 and one water molecule (Figure 4A), and the Ca2+ 276 composed of Asp41, Asn77, Gln2, Ile79, Leu75 and Val81 (Figure 4B). The presence of bound calcium ions is a feature shared by members of the subtilisin superfamily, where calcium binding is shown to be essential for correct folding and structural stability [32]. Considering the stabilizing effect of binding metal ions in many proteins, it would be expected that an increased affinity and the number of bound metal ions should correlate with the thermostability of proteins [39].
In silico study
Molecular docking: In molecular docking using YASARA, the cluster conformation is sorted by binding energy. Negative energies mean no binding while more positive energies indicated stronger binding. Based on the docking prediction result, conclusively, there was a possible binding of substrate to the Rand protease. Blind docking of Rand protease to substrate N-succinylalanyl- alanyl-prolyl-phenylalanine-4-nitroanilide resulted in 25 clusters whereby 4 clusters were observed to involve the catalytic triad of rand protease that is Asp32, His64 and Ser221. This substrate was chosen because it was used in the experiment in the lab for protease assay. So, it should be able to mimic the catalysis properties of Rand protease. Among the 25 clusters, the best-fitted compound complex chosen for further discussion was based on the highest binding energy and smaller dissociation constant Kd. The best clusters show for the value of binding energy and Kd: 7.373 kcal/mol, 3940209.75 pM respectively. Residues involve in binding was Asp32, Ser33, Ser62, His64, Leu96, Thr99, Gly100, Ser101, Ser125, Leu126, Gly127, Ala152, Gly154, Asn155, Tyr217, Asn218, Gly219, Thr220, Ser221, Met222. As shown in (Figure 5), the substrate was docked very near the catalytic triad, Therefore, this docking position is compatible with the predicted catalytic triad.

Molecular Dynamics (MD) simulations: MD simulation is the technique used to analyse the dynamic behavior of the Rand protease and three different organic solvents with different log P values. Energy minimization was done before the MD simulation procedures to remove bad contacts and geometry [40]. This was vital because if an MD simulation was started with bad contracts, the energy in that region would be unrealistically high, and that would either crash the simulation or cause the trajectory to proceed in an unrealistic fashion. MD simulations were carried out in periodic boundary conditions (water molecules) at 298 K, equivalent to 25°C. After a 10 ns trajectory, the resulting outcomes were analysed by means of CRMSD and RMSF.
Root Mean Squared Deviation (RMSD): A common way to analyse the structural stability in MD simulation techniques is to monitor the RMSD of the backbone from the initial structure during the simulation. The RMSD is analysed in order to characterize the conformational changes of the structure. RMSD measures the evolution of the structure’s conformation from the initial conformation [40].
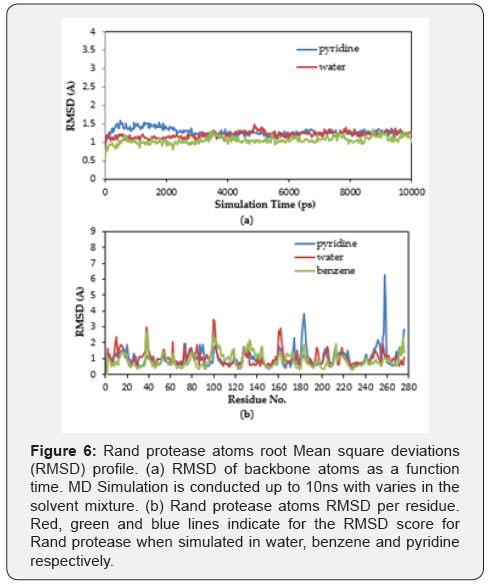
The RMSD of the instantaneous structures of the three models is plotted as a function of time (Figure 6A) and per residue (Figure 6B). The RMSD was evaluated during the simulation, and it is observed that, during the beginning of the trajectory, pyridine conferred a higher RMSD value (1.3Å) compared to the water (1.1Å) and benzene (0.9Å). This suggests that it was increasing in the beginning for pyridine, but after around 3 ns it became almost constant for the rest of the duration of the simulation. This demonstrates that pyridine has a lower RMSD for the Rand protease backbone and has less flexibility, indicating the stable dynamic behavior structure of Rand protease. Furthermore, the RMSD for benzene seems to have reached a plateau early in the simulation, which is after less than 100 ps, and equilibrated around 1.1. Comparative analysis of the average structures at 298 K in water molecules implies that the benzene structure is more stable than that of the pyridine. This finding confirms the previous findings for the wild-type Rand protease [41]. However, the overall RMSD for all proteases is considerably small, indicating that the structure after the simulation is not much different from the initial structure, and the protease structures were fairly capable of keeping their conformation [42].
Root mean square fluctuations (RMSF)
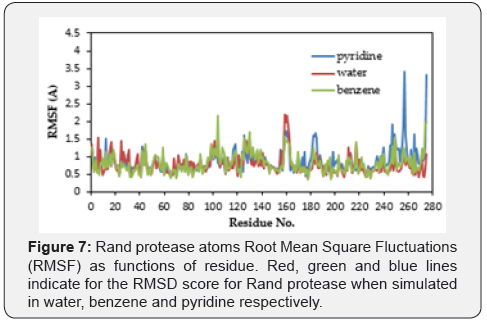
The RMSF is a measurement of the thermal motions of a residue. It captures, for each atom, the fluctuation about its average position. This analysis gives an overview of the flexible regions in the protein and corresponds to the crystallographic b-factors (temperature factors).
The RMSF of the instantaneous structures of the three models is plotted as a function of residue (Figure 7). Briefly, the simulation denotes the stabilization of the Rand protease throughout the simulation, throughout which the overall Rand protease structure does not deviate more than 1.0Å. Superimposition of the 3D structure of the Rand protease before and after simulation also reveals no major changes in terms of the secondary structure.
Conclusion
The protein’s 3D structure is important for designing a novel biocatalyst that suits industrial applications. Therefore, the data from the predicted 3D structure of Rand protease may increase enzyme production and help to analyse and provide an understanding of the structure of the thermostable, organicsolvent- tolerant Rand protease. The Ca2+ is one of the metals that is often identified by researchers to promote the stability of a protein structure, especially a thermostable enzyme. The metals (two Ca2+ atoms) presented in the predicted 3D structure of the Rand protease was suggested to contribute to the stability of the enzyme. It was observed that the simulation in the mixture of benzene and water showed the lowest deviation of the overall Rand protease structure compared to the overall Rand protease structure when simulated in water. This result tallies with the previous Rand protease characterization record, which demonstrates less protease activity in the presence of pyridine as the solvent in the reaction mixture. The thermostable and organic-solvent-tolerant Rand protease is a unique and important enzyme, possessing a novel combination of many industrially important properties, which can tolerate harsh organic solvents and withstand high temperatures. Therefore, Rand protease is a promising candidate for many of the industrial processes in which proteases are utilized.
Acknowledgment
We would also like to express our gratitude to the late Prof. Mahiran Basri, principal investigator in Enzyme and Microbial Technology Research Center, Faculty of Science, University Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia, for sharing her knowledge with us during the course of this research. This project was funded by the Ministry of Science, Technology and Innovation Malaysia (MOSTI) under Malaysian Technology Development Centre (MTDC) for financial aid through the CRDF Bridging Fund (Vote number: 6364703).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Contesini FJ, Melo RR, Sato HH (2018) An overview of Bacillus proteases: from production to application. Crit Rev Biotechnol 38 (3): 321-334.
- Mathew CD, Gunathilaka S (2015) Production, purification and characterization of a thermostable alkaline serine protease from Bacillus lichniformis NMS-1. Int J Biotechnol Mol Biol Res 6 (3): 19-27.
- Bjerga G, Arsın H, Larsen Ø, Puntervoll P, Kleivdal H (2016) A rapid solubility-optimized screening procedure for recombinant subtilisins in E. coli. J Biotechnol 222: 38-46.
- Szilagyi A, Zhang Y (2014) Template-based structure modeling of protein–protein interactions. Curr Opin Struct Biol 24: 10-23.
- Zou T (2014) Protein folding and dynamics using multi-scale computational methods. Thesis, Arizona State University.
- Laane C, Boeren S, Vos K, Veeger C (1987) Rules for optimization of biocatalysis in organic solvents. Biotechnol Bioeng 30 (1): 81-87.
- Bornot A, Etchebest C, De Brevern AG (2011) Predicting protein flexibility through the prediction of local structures. Proteins 79 (3): 839-852.
- James LC, Tawfik DS (2003) Conformational diversity and protein evolution – a 60-year-old hypothesis revisited. Trends Biochem Sci 28 (7): 361-368.
- Markland FS, Smith EL (1971) 16 Subtilisin’s: Primary structure, chemical and physical properties. The Enzymes 3: 561-608.
- Takagi H, Koga M, Katsurada S, Yabuta Y, Shinde U, et al. (2001) Functional analysis of the propeptides of subtilisin E and aqualysin I as intramolecular chaperones. FEBS Lett 508 (2): 210-214.
- Tanaka S-i, Takeuchi Y, Matsumura H, Koga Y, Takano K, et al. (2008) Crystal structure of Tk-subtilisin folded without propeptide: requirement of propeptide for acceleration of folding. FEBS Lett 582 (28): 3875-3878.
- Rai SK, Mukherjee AK (2010) Statistical optimization of production, purification and industrial application of a laundry detergent and organic solvent-stable subtilisin-like serine protease (Alzwiprase) from Bacillus Subtilis DM-04. Biochemical Engineering Journal 48 (2): 173-180.
- Weng M, Deng X, Bao W, Zhu L, Wu J, et al. (2015) Improving the activity of the subtilisin nattokinase by site-directed mutagenesis and molecular dynamics simulation. Biochem Biophys Res Commun 465 (3): 580-586.
- Forrester, Wright J, Forrester JW (1969) Urban dynamics. Ed. mIt press Cambridge.
- Yang L, Dordick JS, Garde S (2004) Hydration of enzyme in nonaqueous media is consistent with solvent dependence of its activity. Biophys J 87 (2): 812-821.
- Krieger E, Koraimann G, Vriend G (2002) Increasing the precision of comparative models with YASARA NOVA-a self-parameterizing force field. Proteins 47 (3): 393-402.
- Krieger E, Darden T, Nabuurs SB, Finkelstein A, Vriend G (2004) Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins 57 (4): 678-683.
- Kuzmanic A, Zagrovic B (2010) Determination of ensemble-average pairwise root mean-square deviation from experimental B-factors. Biophys J 98 (5): 861-871.
- Alvarez M, Zeelen JP, Mainfroid V, Rentier-Delrue Fo, Martial JA, et al. (1998) Triose-phosphate Isomerase (TIM) of the Psychrophilic Bacterium Vibrio marinus Kinetic and structural properties. J Biol Chem 273 (4): 2199-2206.
- Pack SP, Yoo YJ (2004) Protein thermostability: structure-based difference of amino acid between thermophilic and mesophilic proteins. J Biotechnol 111 (3): 269-277.
- Argos P, Rossmann MG, Grau UM, Zuber H, Frank G, eta l. (1979) Thermal stability and protein structure. Biochemistry 18 (25): 5698- 5703.
- Zaki MJ, Bystr off C (2008) Protein structure prediction. (second ed), Humana Press Inc, New York, USA.
- Jain SC, Shinde U, Li Y, Inouye M, Berman HM (1998) The crystal structure of an autoprocessed Ser221Cys-subtilisin E-propeptide complex at 2.0 Ã resolution. J Mol Biol 284 (1): 137-144.
- Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, et al. (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29 (1): 291-325.
- Topf M, Baker ML, Marti-Renom MA, Chiu W, Sali A (2006) Refinement of protein structures by iterative comparative modeling and CryoEM density fitting. J Mol Biol 357 (5): 1655-1668.
- Rawlings ND, Morton FR, Barrett AJ (2006) MEROPS: the peptidase database. Nucleic Acids Res 34 (1): 270-272.
- Reddy L, Wee YJ, Ryu HW (2008) Purification and characterization of an organic solvent and detergent-tolerant novel protease produced by Bacillus sp. RKY3. J Chem Technol Biotechnol 83 (11): 1526-1533.
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography 26 (2): 283-291.
- Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2 (9): 1511-1519.
- Benkert P, Biasini M,Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27 (3): 343-350.
- Siezen R, de Vos W, Leunissen J, Dijkstra B (1991) Homology modelling and protein engineering strategy of subtilases, the family of subtilisinlike serine proteinases. Protein Eng 4 (7): 719-737.
- Siezen RJ, Leunissen JA (1997) Subtilases: the superfamily of subtilisinlike serine proteases. Protein Sci 6 (3): 501-523.
- Kluskens LD, Voorhorst WG, Siezen RJ, Schwerdtfeger RM, Antranikian G, et al. (2002) Molecular characterization of fervidolysin, a subtilisin like serine protease from the thermophilic bacterium Fervidobacterium pennivorans. Extremophiles 6 (3): 185-194.
- Kulakova L, Galkin A, Kurihara T, Yoshimura T, Esaki N (1999) Coldactive serine alkaline protease from the psychrotrophic bacterium Shewanella strain Ac10: gene cloning and enzyme purification and characterization. Appl Environ Microbiol 65 (2): 611-617.
- Yamagata H, Masuzawa T, Nagaoka Y, Ohnishi T, Iwasaki T (1994) Cucumisin, a serine protease from melon fruits, shares structural homology with subtilisin and is generated from a large precursor. J Biol Chem 269 (52): 32725-32731.
- Rawlings ND, Barrett AJ (1999) MEROPS: the peptidase database. Nucleic Acids Res 27 (1): 325-331.
- Hakansson K, Wang AH-J, Miller CG (2000) The structure of aspartyl dipeptidase reveals a unique fold with a Ser-His-Glu catalytic triad. Proceedings of the National Academy of Sciences 97 (26): 14097- 14102.
- Welch AR, McNally LM, Hall M, Gibson W (1993) Herpesvirus proteinase: site-directed mutagenesis used to study maturational, release, and inactivation cleavage sites of precursor and to identify a possible catalytic site serine and histidine. J Virol 67 (12): 7360-7372.
- Betzel C, Teplyakov A, Harutyunyan E, Saenger W, Wilson K (1990) Thermitase and proteinase K: a comparison of the refined threedimensional structures of the native enzymes. Protein Engineering 3 (3): 161-172.
- El-Bastawissy E, Knaggs MH, Gilbert IH (2001) Molecular dynamics simulations of wild-type and point mutation human prion protein at normal and elevated temperature. J Mol Graph Model 20 (2): 145-154.
- Abusham RA, Rahman RNZR, Salleh AB, Basri M (2009) Optimization of physical factors affecting the production of thermo-stable organic solvent-tolerant protease from a newly isolated halo tolerant Bacillus Subtilis strain Rand. Microb Cell Fact 8 (1): 20.
- Gsponer J, Ferrara P, Caflisch A (2001) Flexibility of the murine prion protein and its Asp178Asn mutant investigated by molecular dynamics simulations. J Mol Graph Model 20 (2): 169-182.






























