Prevalence of Virulance Gene by Real Time Pcr of Legionella pneumophila Strains Isolated from Different Sources
Gülnur Tarhan1*, Sadık Akgün1, Selin Nar Ötgün2, Meral Karaoğlan2 and Yusuf Özay 3
1Department of MedicalMicrobiology, Adıyaman University,Faculty of Medicine, Turkey
2RespiratoryPathogens Reference Laboratory, National PublicHealthInstitute, Turkey
3Department of Medical Biology, Adıyaman University,Faculty of Medicine, Turkey
Submission: September 01, 2018; Published: October 17, 2018
*Corresponding author: GülnurTarhan, Department of MedicalMicrobiology, Adıyaman University,Faculty of Medicine, Turkey, Tel: 905059448568; Email: gulnur.tarhan@yahoo.com
How to cite this article: Gülnur T, Sadık A, Selin N Ö, Meral K, Yusuf Ö. Gülnur Tarhan1*, Sadık Akgün1, Selin Nar Ötgün2, Meral Karaoğlan2 and Yusuf 002 Ö. Adv Biotech & Micro. 2018; 11(5): 555821. DOI:10.19080/AIBM.2018.11.555821
Abstract
Legionella pneumophila is a major ethiological agent of legionnaires’ disease and Pontiac fever which is an important lung infection on old patients that smoke more than a pack of cigarettes a day and are addicted to alcohol or those whose immunity system have been pressurized. For the formation of disease, numbers of bacteria in inhaled aerosol are important as well as pathogenity degree of bacteria which means virulence. The virulence of this bacteria can change depending on chemical, physical and biological conditions of the environment where they live. The entire infection process of bacteria is linked to virulence factors such as bacterial cell attachment to host cells, survival and intracellular replication and cell-to-cell, quarum sensing encoded by virulence genes.
The products of genes involved in the initial attachment to host cells and early stages of intracellular infection include type IV pili, the 60 kDa heat-shock protein Hsp60, the poreformation protein RtxA, the macrophage infectivity potentiator Mip and the macrophage-specific infectivity protein MilA and quorum sensing. The aim of this study was to investigate macrophage infectivity potentiator gene, quarum sensing and pore formation protein gene that played important roles in various steps of the whole infection process in 15 enviromental and 10 clinical L. pneumophila isolates. All clinical isolates were positive for the Legionella quorum sensing cluster. Nine clinical isolates were positive for Mip gene region. RtxA gene locus was positive 8 clinical isolates. All of Mip, RtxA, Lqs and HdeD gene region were positive in 10 environmental isolates. While L. pneumophila serogroup 1 standard strain ATCC33152 strain waspotisitve for all gene region, only Lqs region was negative in L. pneumophila serogroup 1 standard strain ATCC43111 strain. Legionella micdadei was found positive for all gene region. While Mip gene region was negative, other gen regions were positive for L.bozemanii.
Keywords: Leigonella; virulance gene; Mip; RtxA; Lqs; HdeD
Introduction
Legionella is an intracellular pathogen that parasites host mononuclear phagocytes. There have nearly 59 species and 70 distinct antigenic types of Legionella. Of the 50 species, 20 are disease-causing agents [1,2]. Between the these species; Legionella pneumophila is an ethiological agent of Legionnaires’ disease and the most common species detected in clinical cases (80-90%), other species have also been reported to cause disease. It consists of 16 serogroups [3,4]. The majority of isolates in the CDC collection are serogroup 1 (sg 1), which are the primary cause of outbreaks (76.5% to 90%). Other Legionella species have also been reported to cause disease. The most common species after L. pneumophila is L. micdadei [5].
They are located water reservoirs, natural water sources such as lakes and rivers or in the biofilm bed or in the free water phase in artificial water environments such as ventilation systems, humidifiers, cooling kettles, spa centers, decorative water fountains, jacuzzi and shower heads, ice machines, vegetable humidifiers and dental units [6-8]. L. pneumophila can be transmitted to a human host by inhalation of aerosolized water from a contaminated man made water system. However, little is known about Legionnaires’ disease caused by these non-pneumophila species and their specific properties [9,10]. Legionella bacteria have become a microorganism since the first isolates, in order to detect the existence of these bacteria and to develop methods of fighting and to investigate pathogenic mechanisms. However, the presence of the bacteria in water systems does not always cause outbreaks. Whether there is an outbreak depends on various factors, such as the diversity of serogroups, inhaled bacteria count, differences in immunity in the population, and the virulence of the bacteria [11,12]. Therefore; although preventing Legionella colonization in water systems is the priority, determining a type IV secretion system (T4SS) their virulence is also very important.
The virulence factors characterized include genes required for the whole infection process, such as bacterial cell attachment to host cells, survival and intracellular replication and cell-to-cell spread. The products of genes involved in the initial attachment to host cells and early stages of intracellular infection include type IV pili, the 60 kDa heat-shock protein Hsp60, the poreformation protein RtxA, the macrophage infectivity potentiator Mip and the macrophage-specific infectivity protein MilA [13-16]. At the same time, L. pneumophila harbors the Legionella quorum sensing cluster (Lqs ), which includes genes encoding the autoinducer synthase LqsA, the sensor kinase LqsS, the response regulator LqsR, and a homologue of HdeD, which is involved in acid resistance in Escherichia coli. LqsR promotes host-cell interactions as an element of the stationary-phase virulence regulatory network [17-20].The aim of this study was to investigate macrophage infectivity potentiator gene, quarum sensing and pore formation protein gene in 15 enviromental and 10 clinical L. pneumophila isolates.
Materials and Methods
All consumables used in this study were obtained from Adıyaman University Scientific Research Projects Unit Project No: TIPFMAP / 2015-0006.
Bacterial isolates
10 clinical L. pneumophila isolates, two L. pneumophila serogroup 1 standard strain (ATCC 43111, ATCC 33152), 1 Legionella bozemanii (ATCC 33217) and 1 Legionella micdadei standard strains (ATCC 33218) were obtained from collections at the National Public Health Institute, Respiratory Pathogens Reference Laboratory in Ankara, Turkey. 15 enviromental L. pneumophila were isolated from water systems.
Culturing of bacterial isolates
All isolates to be used in the study were inoculated on the BCYE Agar medium by the reduction method. All inoculums were incubated at 37 °C and 5% CO2 for 3-4 days. At the end of this period, the colonies were stained with Gram stain method and the morphological characteristics of the bacteria were identified by light microscopy.
DNA extraction
A loopful of cells were suspended in 500 μl of TE buffer (10mM Tris , 1mM EDTA , pH 8.0 ) in a 1.5ml screw- cap microcentrifuge tube. The suspension was centrifuged at 15,000xg for 10min. The supernatant was removed and then the pellet was washed twice with 500 μl of TE buffer and then resuspended with 200μl of the same buffer. The samples were incubated in a boiling water bath for 20 min centrifuged, and supernatants containing DNA was transferred to clean microcentrifuge tubes and kept at - 20 °C until used [21].
Real time PCR reaction
Three virulence gene region was examined to evaluate type II secretion system, type IV secretion system and quarum sensing cluster including Mip, RtxA, Lqs and HdeD (15,17,22). Primer sequences for each gene region were shown in Table 1.
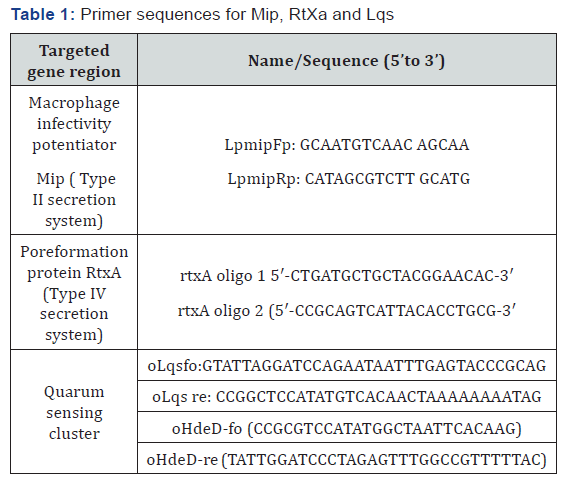
The 25 μl reaction mixture contained 12.5 μl of Lightcycler FastStart Reaction mix (including FastStart Taq DNA polymerase with buffer, dNTP mix, SYBR Green I dye, and 10 mM MgCl2), additional MgCl2 to achieve the optimal final concentration of 4 mM, each primer at 0.5 mM, 1 U of uracil- DNA-glycosylase (UNG), and 5 μl of template DNA. Before amplification, the capillaries were kept at room temperature for 10 min to allow UNG to break down the possible contaminating amplicons and then were heated to 95 °C and held at that temperature for 10 min to deactivate UNG and activate the polymerase enzyme prior to the start of cycling. Amplification and steps were standardized according to reference literatures [15,17,22].
Reactions with purified L. pneumophila DNA at three concentrations (100, 20, and 4 ng per capillary) were included in each run to construct the standard curve. Quantification was performed according to the instrument manual by setting the noise band over the background fluorescence and determining the crossing points arithmetically with the use of two fit points. Melting points were calculated by the instrument. In order to normalize the run-to-run variations in the measured melting points, a melting temperature (Tm) ratio was calculated by dividing the melting point of each sample by the melting point of the 20-ng standard sample.
Results
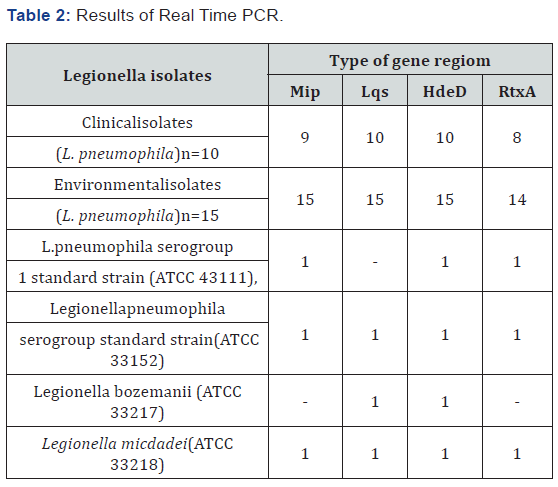
A total of 25 L. pneumophila isolate (10 clinical and 15 environmental ) , 1 standard L. pneumophila serogroup 1 (ATCC 43111), 1 Legionella pneumophila serogroup 1 standard strain(ATCC 33152), Legionella bozemanii(ATCC 33217) and Legionella micdadei(ATCC 33218) standard strain was tested in this study. All samples were examined by Real Time PCR for type II secretion system, type IV secretion system and quarum sensing cluster using Mip, RtxA, Lqs and HdeD gene region. All clinical isolates were positive for the Legionella quorum sensing (lqs) cluster. Of 10 clinical isolates; 9 was positive for Mip gene region. This gene locus was not detected in 1 isolate. RtxA gene locus was positive 8 clinical isolates. Of 10 environmental isoltes All of Mip, RtxA, Lqs and HdeD gene region were positive in ten environmental isolates. While L. pneumophila serogroup 1 standard strain ATCC33152 strain waspotisitve for all gene region, only Lqs region was negative in L. pneumophila serogroup 1 standard strain ATCC43111 strain. Legionella micdadei was found positive for all gene region. While Mip gene region was negative, other gen regions were positive for L.bozemanii (Table 2).
Discussion
Legionella pneumophila is the ethiological agent of Legionellosis. About 90% cases of this disease are due to this species, and the predominant serogroup (sg) 1 of L. pneumophila accounts for 84% of cases (1). The severity of bacterial disease depends on the virulence properties of microorganism. L.pneumophila has a many of the traditional bacterial determinants that are important for pathogenicity in other bacteria, such as lipopolysaccharide, flagella, pili, T2SS, T4SS and outer membrane proteins. T4SS translocates around 200 effector proteins, including many proteins with eukaryotic similarity, into the host cell, where they act on diverse host cell pathway [19-21]. Among these region, L. pneumophila virulenceassociated response regulator was directly promote pathogen host cell interactions, such as phagocytosis, formation of the LCV, intracellular replication, and cytotoxicity, while delaying the entry of L. pneumophila into the replicative growth phase. These virulence factors characterized include genes required for the whole infection process, such as bacterial cell attachment to host cells, survival and intracellular replication and cell-to-cell spread.
The products of genes involved in the initial attachment to host cells and early stages of intracellular infection include type IV pili, the 60 kDa heat-shock protein Hsp60, the poreformation protein RtxA, the macrophage infectivity potentiator Mip and the macrophage-specific infectivity-protein MilA and quarum sensing interactions [12-15]. The genes required for bacterial survival and intracellular replication are a group of genes called icm (intracellular multiplication) . The L. pneumophila virulenceassociated response regulator is located within the Lqs cluster (lqsA-lqsR-hdeD-lqsS). This cluster includes genes encoding the autoinducer synthase LqsA, the sensor kinase LqsS, and the putative membrane protein HdeD. LqsR was directly promote pathogen host cell interactions, such as phagocytosis, formation of the LCV, intracellular replication, and cytotoxicity, while delaying the entry of L. pneumophila into the replicative growth phase. The clustering and orientation of the lqsA-lqsR-lqsS genes are conserved among different bacterial species [19,20].
The Lqs cluster harbors four genes (lqsA-lqsR-hdeD-lqsS) and is present in all L. pneumophila strains. The purpose of this study was to investigate macrophage infectivity potentiator gene, quarum sensing and pore formation protein gene that played important roles in various steps of the whole infection process in 15 enviromental and 10 clinical L. pneumophila isolates. All samples were examined by Real Time PCR for type II secretion system, type II secretion system and quarum sensingcluster using Mip, RtxA, Lqs and HdeD gene region. All clinical and environmental isolates were positive for the Lqs cluster. Lqs gene region in L. pneumophila serogroup 1 standard strain (ATCC 43111) was not detected. There are some published data for the detection of Mip, RtxA gene in clinical and environmental samples. Mip and RtxA genes are commonly detected in clinical and environmental isolates. The results obtained from these gene regions were found to be consistent with the study done by Huang et al [5]. In the Legionella bozemanii standard strain, Mip and RTxA gene region was negative. All gene regions were positive in the case of the Legionella micdadei standard strain evaluated simultaneously in the study. There is no study of the cluster of Lqs in clinical and environmental samples. This study is the first study of a cluster of Lqs in clinical and environmental samples [5,11,20,22].
Conclusions
In our study Mip, RtxA and Lqs cluster genes were commonly detected in clinical and environmental samples. These gene regions can be used directly from clinical and environmental samples of L. pneumophila. In order to evaluate the pathogenicity factors encoded by these genes, there is a need for detailed studies involving enzymatic, cellular and protein activities.
Conflicts of interest
The authors declare that they have no competing interests.
Acknowledgements
This study was supported by the Scientific Research Projects Unit of Adıyaman University (Project number: TIPFMAP / 2015- 0006).
References
- Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, et al. (2002) Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J Infect Dis 186(1): 127-128.
- Kozak NA, Benson RF, Brown E (2009) Distribution of lag-1 alleles and sequence-based types among Legionella pneumophila serogroup 1 clinical and environmental isolates in the United States. J Clin Microbiol 47(8): 2525-2535.
- Benin AL, Benson RF, Besser RE (2002) Trends in legionnaires disease, 1980-1998: declining mortality and new patterns of diagnosis. Clin Infect Dis 35(9): 1039-1346.
- McNally C, Hackman B, Fields BS, Plouffe JF (2000) Potential importance of Legionella species as etiologies in community acquired pneumonia (CAP). Diagn Microbiol Infect Dis 38(2): 79-82.
- Huang B, Yuan Z, Heron BA, Gray BR, Eglezos S, et al. (2006) Distribution of 19 major virulence genes in Legionella pneumophila serogroup 1 isolates from patients and water in Queensland, Australia. J Med Microbiol 55(8): 993-997.
- Atlas RM (1999) Legionella: from environmental habitats to disease pathology, detection and control. Environ Microbiol 1(4): 283-293.
- Lau HY, Ashbolt NJ (2009) The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water. J Appl Microbiol 107(2): 368-378.
- Benowitz I, Fitzhenry R, Boyd C, Dickinson M, Levy M, et al. (2018) Rapid Identification of a Cooling Tower-Associated Legionnaires’ Disease Outbreak Supported by Polymerase Chain Reaction Testing of Environmental Samples, New York City, 2014-2015. J Environ Health 80(8): 8-12.
- Hamilton KA, Hamilton MT, Johnson W, Jjemba P, Bukhari Z, et al. (2018) Health risks from exposure to Legionella in reclaimed water aerosols: Toilet flushing, spray irrigation, and cooling towers. Water Research 134: 261-279.
- Oliva G, Sahr T, Buchrieser C (2018) The Life Cycle of L. pneumophila: Cellular Differentiation Is Linked to Virulence and Metabolism. Front Cell Infect Microbiol 8: 3.
- Zhan XY, Zhu QY (2017) Molecular evolution of virulence genes and non-virulence genes in clinical, natural and artificial environmental Legionella pneumophila isolates. Peer J 5: e4114.
- Vogel JP, Andrews HL, Wong SK, Isberg RR (1998) Conjugative transfer by the virulence system of Legionella pneumophila. Science 279(5352): 873-876.
- Cianciotto NP, Fields BS (1992) Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc Natl Acad Sci 89(11): 5188-5191.
- Cirillo SLG, Lum, J, Cirillo, JD (2000) Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146: 1345-1359.
- Cirillo SLG, Yan L, Littman M, Samrakandi MM, Cirillo JD (2002) Role of the Legionella pneumophila RtxA gene in amoebae. Microbiology 148(6): 1667-1677.
- Harb OS, Abu Kwaik Y (2000) Characterization of a macrophagespecific infectivity locus (milA) of Legionella pneumophila. Infect Immun 68(1): 368-376.
- Tiaden A, Spirig T, Carranza P, Brüggemann H, Riedel K, et al. (2008) Synergistic Contribution of the Legionella pneumophila lqs Genes to Pathogen-Host Interactions. J Bacteriol 190(22): 7532-7547.
- Tiaden A, Spirig T, Sahr T, Wälti MA, Boucke K, et al. (2010) The autoinducer synthase LqsA and putative sensor kinase LqsS regulate phagocyte interactions, extracellular filaments and a genomic island of Legionella pneumophila. Environ Microbiol 12(5): 1243-59.
- Tiaden A, Spirig T, Weber SS, Brüggemann H, Bosshard R, et al. (2007) The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell Microbiol 9 (12): 2903-2920.
- Personnic N, Striednig B, Hilbi H (2018) Legionella quorum sensing and its role in pathogen-host interactions. Curr Opin Microbiol 41:29- 35.
- Kocagöz T, Yılmaz E, Özkara Ş, Kocagöz S, Hayran M, et al. (1993) Detection of Mycobacterium tuberculosis in sputum samples by polymerase chain reaction using a simplified procedure. J Clin Microbiol 31(6): 1435-1438.
- Deborah AW, Lieberman BY, Reischl U, Gordon SM, Procop GW (2003) Detection of Legionella pneumophila by Real-Time PCR for the mip Gene. J Clin Microbiol 41(7): 3327-3330.






























