Role of Rumen Protozoa: Metabolic and Fibrolytic
Shreya Patel* and Padma Ambalam
Department of Biosciences, Saurashtra University, India
Submission: December 27, 2018; Published: August 31, 2018
*Corresponding author: Shreya Patel, Department of Biosciences, Saurashtra University, Rajkot- 360005, Gujarat, India, Tel: 096246823671; Email: shreya29patel@gmail.com
How to cite this article: Shreya Patel, Padma Ambalam. Role of Rumen Protozoa: Metabolic and Fibrolytic. Adv Biotech & Micro. 2018; 10(4): 555793. DOI:10.19080/AIBM.2018.10.555793
Abstract
Ruminant animals are having a diverse microbial ecosystem for digesting fibrous feeds and protozoa can represent about half of the total rumen microbial biomass. To explain the role of ruminal protozoa, defaunated and refaunated animals were compared from decades. It is the fact that protozoa are useful for numerous reasons to their host. From those, first is metabolism and the other is fiber digestion. Protozoa play a key role in maintaining fermented product concentration like methane, ammonia, lactate, propionate, butyrate, etc. Plant cell walls are complex structure and yet there is not complete understanding of it. Rumen microbes have developed an efficient strategy to digest this complex structure to make final products available to the host. However, digestion of feeds remains incomplete, and numerous research attempts have been made to increase this extent of digestion. This mini review briefly describes the role of protozoa in ruminants’ metabolism and fiber degradation.
Keywords:Rumen; Protozoa; Fiber degradation; Morphology
Abbrevations:VFA: Volatile Fatty Acids; EST: Expressed Sequence Tag; CMC: CarboxyMethyl Cellulose
Introduction
Rumen is first chamber of the gastrointestinal track of ruminant where digestion of the plant fibres is taken place and solids are absorbed followed by reticulum site for waste removal. After reticulum, omasum and abomasum are like other non-ruminants followed by small intestine. After a long time period from the first exposure to rumen, researchers started taking interest in rumen protozoa. After 1920, significant studies were started in identification, morphology and functions of rumen protozoa [1]. In addition to that it was observed that presence and absence of particular rumen protozoa affect factors like pH, ammonia concentration and also type and number of bacteria and these all factors affect digestion in ruminants [2]. Active participation of ciliate protozoa in digestion process of ruminants was earlier suggested by Tier and Hungate and Coleman et al., [3] and then was proven earlier by in vivo and in vitro studies [4-6] and also further supported till date by various studies relating to that [7-9].
Followed by most abundant bacteria, ciliate protozoa are the second most populated organisms in bovine rumen [10]. These ciliate protozoa are classified into mainly two groups viz. holotrichs and entodinomorphs (oligotrichs) based on their morphological characteristics. Also, they can be classified as soluble sugar utilizers, starch degraders and lignocellulose hydrolysers on the basis of their substrates. Larger protozoa generally ingest plant structural polymer and smaller one prefers storage polymers and sugars [8]. The holotrichs use soluble sugars and convert them rapidly into stored polysaccharide which is used during periods when sugars are not available and if provided with excess sugar they store starch until they burst [9]. holotrichs are having cilia over their entire body and generally sugar utilizers while entodinomorphs are having cilia over discreet region and found to be attached to fibre, utilize starch as well as various plant materials [11-12]. Among these, the holotrich protozoa are represented by 15 different genera in the rumen of different animals and genera include Isotricha, Dasytricha, Buetschlia and Charoninaf which are widely distributed in the rumen of domestic and wild ruminants and hind gut fermentors. Entodinomorphs are represented by several abundant genera viz. Entodinium, Diplodinium, Eremoplastron, Eudiplodinium, Elytroplastron, Metadinium, Ostracodinium, Epidinium. Though, the composition of rumen ciliates varies by geographic regions, host species and their feed consumption. A large number of ciliates have been observed in different conditions but the count for a specific animal is 30 or fewer [10].
Role of Protozoa in Digestion
As discussed earlier, ruminants are herbivorous mammals having specific organ called rumen and function of rumen mainly includes conversion of feed materials into the form which can be easily utilized by animals as a source of energy. Ruminants are able to digest plant material like cellulose, xylan, starch, sucrose etc. through diversified microbes present in it. Ingested feed is converted into volatile fatty acids in this rumen. Anaerobiosis is maintained by gases generated by fermentation viz. carbon dioxide, methane and traces of hydrogen. However, some amount of oxygen is entered in rumen which is utilized by some facultative anaerobes and anaerobic condition is maintained. As reported, rumen is having highly diversified microbial ecosystem. Anaerobic bacteria are most abundant followed by ciliate protozoa, anaerobic fungi and bacteriophages. Among this highly diversified microbial system some are dependent on others for nutrient supply required for them. But each of these microbes has significant fibrolytic activity. Among these fibrolytic microbes, protozoa contribute 19-28% of cellulase activity of total rumen fibrolytic activity [11]. Apart from fibrolytic activity, rumen habituated protozoa are a good source of lipid as well and up to 27% from the total lipids is thought to be from holotrichs. Also, rumen protozoa are having much concentration of unsaturated fatty acids than other rumen parasites. Thus, they help to lower the sum of fully saturated fatty acids by incorporating them into their membrane phospholipids. Protozoa protein contains glutamic acids, leucine, lysine, and isoleucine as their backbone amino acids make their protein easily digestible than other rumen microbial protein [13].
Protozoa and rumen metabolism
There are several ways to analyse the role of ciliates in rumen metabolism. First is defaunation and refaunation in which animals are defuanated i.e. protozoa free animals are tested for studying the role of rumen function without changing the regular diet of an animal. Also, there are reports on defaunation and after that the animal was inoculated with only one species i.e. monofaunated to check the role of respective protozoan species [12]. There was a report of meta-analysis which was conducted to study the main effects of defaunation based on 23 in vivo studies comprising 48 comparisons [14]. For defaunation purpose, sodium lauryl sulfate, alkanes, synperonic NP9, calcium peroxide, copper sulfate etc. chemicals are used [15]. Apart from this method, second is rumen manipulation and third is isolation of newborn from their mother were used to defaunate the animal. Rumen manipulation is carried out by empting and subsequent washing the rumen by chemicals. In third method, newborns are separated from mother and other animals and are kept in isolated place [16]. There is a direct correlation observed between protozoa concentration and quantity of fermentation products. Whatever feed ingested by the ruminant, is enzymatically converted into VFAs (Volatile Fatty Acids) that plays a pivotal role in providing an animal their basic nutrition (http://www.fao.org). Dietary carbohydrates which are cellulose, hemicellulose, starch, soluble sugars and pectin, are degraded to acetate, propionate and butyrate like principal VFAs (Figure 1) [17-18]. The production of fermentation products is observed to be different in faunated and defaunated animals. Methane and ammonia, these two gases are mainly involved fermented products of metabolic activity by protozoa. As protozoa concentration decreases, bacterial degradation by protozoa simultaneously decrease resulted in decreasing ammonia which is probably the most consistent results observed due to defaunation. As the size of protozoa increases, its predation activity increases. According to that larger entodinomorphids having greater predatory activity than smaller holotrichs as predatory activity is fully dependent on the size of the protozoa. This ultimately shows a lower impact on ammonia concentration and protein flow of duodenal. In the case of methane, protozoa directly don’t produce methane, but they enhance the growth of methanogenic bacteria and consequently enhance methane production [14,19]. Apart from ammonia and methane, defaunation non-significantly affect lactate concentration. There is a higher lactate level report in defaunated animal as protozoa grab lactate more rapidly than bacteria, but this increased lactate level i.e. about 11% was nonsignificant [20].
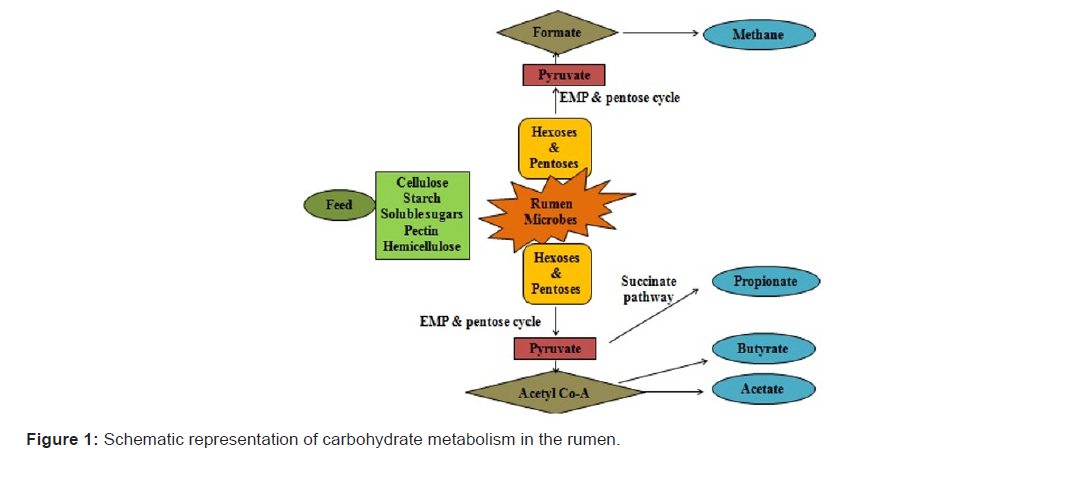
Shifting from lactate concentration towards rumen pH, the rumen pH is shown increased after defaunation with diets containing a high level of concentrate, whereas it is reported to be decreased with diets containing a low level of concentrate. VFA concentration was observed to be decreased i.e. about 5%. Decrease in VFA level clearly highlights the role of these ciliates in rumen metabolism and synthesis of VFA. Protozoa engulf exogenous fatty acids and divert more carbon toward VFA production for fatty acid synthesis and simultaneously increase VFA production. Protozoa has noteworthy role in butyrate and acetate production and bacterial butyrate and acetate production can’t compensate this much quantity after defaunation. This fall in concentration may be derived either because of inability to reach the level without protozoa or decrease in bacterial activity. However, propionate production by these ciliates is pretty less in quantity. So, credit of propionate production in rumen clearly goes to rumen bacteria. Moreover, increase in propionate concentration after defaunation is observed which may be due to increase in propionate producing bacteria after defaunaion. Along with these acids, VFA concentration is found to be decreased which shows that acid contribution to VFA production by protozoa is quite noteworthy. In addition to that, defaunation is generally correlated with decrease in organic matter digestion and this decrease enhances ultimately VFA production [21-22].
As observed, protozoa especially holotrichs have a strong preference for the starch. But they on the other side inhibit the growth of the starch digesting bacteria as there is a strong nutritional competition between protozoa and these bacteria. In addition to this nutritional competition, protozoa also selectively ingest these starch digesting bacteria attach on the starch granule. These starch digesting bacteria produce propionate, succinate and lactate. Succinate and lactate are further catabolized to propionate by other rumen bacteria. And so, decrease in these group of bacteria consequently decrease propionate production. Therefore, defaunation ultimately increase the production of these bacterial fermented products viz lactate, propionate and succinate. Along with these metabolic products, defaunated animals have significantly higher numbers of ruminal bacteria than control animals and the reason simply seem to be based on environmental niche for the bacteria when protozoa are’t there in the rumen [23].
Apart from effect of defaunation on metabolic products, absence of protozoa may affect rumen pathogenicity. As protozoa engulf pathogenic bacteria and lower the pathogenicity, its absence may raise rumen pathgogenicity. On the other side, defaunation may decrease pathogenicity depends on protozoa population. Hence more research is needed in this area of effect of defaunaation on rumen pathogenicity [24].
Fibrolytic activity of rumen protozoa
As reported till date, rumen protozoa are seizing significant enzymatic activities which contribute imperative role in digestion of ruminants. Enzymatic activity can be studied using diverse strategies viz. direct biochemical detection of specific enzymes from extract of mixed rumen protozoa or from culture of single protozoa, gene cloning of genes encoding specific enzymes and recently by sequencing of protozoan-derived expressed sequence tag (EST) libraries [25-26]. Metagenomic approach is reported mainly in microbial and metabolic functional study of rumen protozoa as difficulty of maintaining these organisms in axenic cultures and which avoiding the difficult culturing step [26]. As known protozoa are known for its fibre degradaing activity in the hosts and this fibrolytic activity had been determined and characterized using varied strategies as discussed previously like direct biochemical detection of specific enzymes in the protozoa derived extract. Another way is by molecular cloning to identify enzyme encoding genes directly and the most recent I.e. to sequence EST libraries. Early reports are there proving fibrolytic activities of these rumen protozoa like Howard et.al established the capacity of protozoan species to express their own enzymes for degradation of plant material, especially Epidinium caudatum was observed to be having this acivity. Similarly, E. ecaudatum had been identified for both its cellulase and hemicellulase activities. Clayet et al., [27] identified 10 different enzyme activities for plant cell wall degradation and their catalytic activities include glycoside hydrolases.
Degradation of cellulose and hemicellulose
Rumen protozoa are having a range of fibrolytic activity involving glycoside hydrolases, and polysaccharide depolymerases which degrade polysachharides that is forming plant cell wall structure. A large number of studies of fibrolytic activity have been done on predominant hydrolytic bacteria as role of rumen ciliates in the degradation of plant cell wall polymers was controversial earlier. Various fibrolytic activities were also observed in rumen ciliates then after from single cell as well as in mixed culture [28-29]. It was believed earlier that cellulolytic activity of rumen ciliate is due to their intracellular bacteria. But afterwards it was proven that it could be both from cellullase activity of engulfed bacteria and also cellulase production of ciliates themselves. Coleman in 1987 reported cellulose activity of rumen ciliate, Eudiplodinium magii using sterile Medium and 14C- labelled plant saccharides. Medium was defaunated and refaunated with Eudiplodinium magii and 14C- labelled cellulose, starch, glucose was used to track conversion of cellulose. The rate of release of soluble 14C-labelled compounds from phosphoric acid-regenerated 14C cellulose by protozoal broken-cells was measured. In that experiment, Coleman observed that 70% of the cellulose was digested in 48h and had an activity of 0.55 to 3.3μg cellulose digested h-l (mg protein)-l. Cellulase activity of protozoa was further confirmed by measuring activity with and without antibiotics as in the presence of antibiotics bacterial cellulose activity was decreased [30]. Role of ciliates in total fiber digestion and in cellulose digestion was also checked by Coleman et al. in 1987 using artificial culture. Cellulase activity was checked using substrate CMC (CarboxyMethyl Cellulose) and change in activity was measured by altering parameters like dilution rate and loading rate of grass substrate. CMCase activity in faunated and defaunated rumen culture using filter paper cellulose as substrate revealed that after defaunation, activity was decreased. At the end, it was concluded that the ciliates contributed about 19-28% in CMCase activity [31]. Defaunation of rumen ciliates has been showing decrease in cellulose and hemicelluloses degradation of about 20% in sheep in one study conducted by Jouany and Ushida in 1994 [23]. Endoglucanase and xylanase activities of three rumen protozoa, Polyplastron multivesiculatum, Eudiplodinium maggii, and Entodinium sp. were compared qualitatively by zymograms and quantitatively by measuring specific activities against different polysaccharides. As reported previously, sheep were defaunated and then refuanated with 4-5 cells of one of above three protozoa. Sonicated protozoal suspension was used as a crude protein extract and protein concentration was measured using Bradford assay using BSA as a standard and CMC was used as substrate for CMCase specific activity. P. multivesiculatum and E. Maggii belonging to large Ophryoscolecidae showed much higher specific activities than the small Ophryoscolecidae and.... Entodinium species. Multiple endoglucanases activities were identified from above mentioned two large Ophryoscolecidae in parallel studies [32-33]. Metagenomics has been a promising approach and fastest advancing field in modern microbiology and more advantageous as >99% microbes from the environment can resist cultivation in laboratory. Metagenomic study of rumen microbes leads to a promising approach to identify novel enzymes with substrate specification including cellulose, hemicellulose, starch, kitin, peptides, etc. [34-37].
Degradation of starch
Coleman reported the amylolytic activity from 14 rumen ciliate species individually by preparing cytoplasmic fractions from those species. He also reported this activity from two natural mixed populations from sheep rumen in 1986. Eremoplastron bovis is observed to be having highest activity for amylase followed by Diploplastron affine, Ophryoscolex caudatus and Polyplastron multivesiculatum grown in vitro on grass and whole meal flour. While Ostracodinium obtusum bilobum and Diplodinium pentacanthum are found to be having lowest activity. Almost all rumen protozoa are having amylase activity. However, concentration of amylase varies between species to species and found to be more than 20fold. Another enzyme maltase is also represented in all species which is less active than amylase [38-39]. In bovine rumen, amylolytic activity has not been well studied and till date only few reports on amylolytic activity of bovine rumen protozoa. In 2012, it was checked in one of the entodinomorphs which is Diploplastron affine from sheep rumen. Protozoal cell extract degrade starch to reducing products with the rate being equivalent to 2.4± 0.47μmol/L glucose per mg protein per min [40]. Similarly, from Eudiplodinium magi, amylolytic activity was checked previously in 2007. The crude enzyme preparation obtained from the bacteria-free ciliates degraded starch at a rate of 29.5 and dextrin at 19.4μmol released glucose/mg protein/h, respectively. These enzymes degraded starch into mainly maltose and maltotriose. Starch degrading numerous fractions is collected upon ion-exchange chromatography of a crude enzyme preparation. Partial sequences from two genes coding for synthesis of α-mylase enzymes were identified in a cDNA library of Eudiplodinium maggii. Gene 1 was of 1625bp length and the gene 2 of 1593 bp. They encoded enzymes of 505 and 431 amino acids, respectively and α-amylase in nature [41]. Ciliates engulf starch grains into vesicles and for some ciliates it takes few seconds to be completely filled with starch grains and some take few hours. It takes approximately 36hours to completely metabolize engulfed starch grains and if it is not given, they are died off. Some species can incorporate this engulfed starch in protein metabolism and in this case at least some part is mediated via intracellular bacteria [39].
Degradation of xylan
Large entodinomorphs are able to digest plant particles. Only Polyplastron and Eudiplodinium are found to digest xylan. Epidinium is insufficient in digesting xylan and also holotrichs have no activity against it [25,37].
Conclusion
Rumen protozoa are key organisms along with bacteria and bacteriophages in this anaerobic chamber. Fibrolytic activity of these difficult to cultivate organisms was studied previously. However, their specific growth requirement in liquid medium has limited understanding of their fibrolytic activity. Still many studies have proved role of these organisms in rumen metabolism and enzymatic activities. These ciliates are having significant role in rumen metabolism though they can increase methane production which is not eco-friendly in excess amount. Key fibrolytic activities observed in these ciliates are against starch, cellulose and hemicelluloses.
References
- Williams AG, Coleman GS (1997) The Rumen Protozoa. In Hobson PN & Stewart CS (Eds.): Springer, Netherlands, pp. 73-139.
- Demeyer DI (1981) Rumen microbes and digestion of plant cell wall. Agriculture and Environment 6(2-3): 295-337.
- Coleman GS (1998) Rumen ciliate protozoa. Advances in parasitology 18: 121-173.
- Akin DE, Amos HE (1979) Mode of attack on orchard grass leaf blades by rumen protozoa. Appl Environ Microbiol 37(2): 322-338.
- Amos HE, Akin DE (1978) Rumen protozoal degradation of structurally intact forage tissues. Appl Environmen Microbiol 36(3): 513-522.
- Yodel RD, Trenkle A, Burroughs W (1966) Influence of rumen protozoa and bacteria upon cellulose digestion in vitro. J Anim Sci 25(3): 609- 612.
- Orpin CG (1983) The role of ciliate protozoa and fungi in the rumen digestion of plant cell walls. Animal Feed Science and Technology 10(2-3): 121-143.
- Stern MD, Hoover WH, Leonard JB (1977) Ultrastructure of rumen holotrichs by electron microscopy. Journal of dairy science 60(6): 911- 918.
- Williams AG (1986) Rumen holotrich ciliate protozoa. Microbiol Rev 50(1): 25-49.
- Kamra DN (2005) Rumen microbial ecosystem. Current science 89(1): 124-135.
- Takenaka A, Tajima K, Mitsumori M, Kajikawa H (2004) Fiber digestion by rumen ciliate protozoa. Microbes and Environments 19(3): 203- 210.
- Williams AG, Coleman GS (1992) The Rumen Protozoa: New York: Springer-Verlag, USA.
- Harfoot CG, Hazlewood GP (1997) Lipid metabolism in the rumen: Springer, Dordrecht, Netherlands.
- Kreuzer M, Kirchgessner M, Müller HL (1986) Effect of defaunation on the loss of energy in wethers fed different quantities of cellulose and normal or steamflaked maize starch. Animal Feed Science and Technology 16(3): 233-241.
- Santra A, Karim SA (2003) Rumen Manipulation to Improve Animal Productivity. Asian-Australian journal of Animal Sciences 16(5): 748- 763.
- Fonty G, Jouany JP, Senaud J, Gouet P, Grain J (1984) The evolution of microflora, microfauna and digestion in the rumen of lambs from birth to four months. Canadian Journal of Animal Sciences 64(5): 165-169.
- Bergman EN (1990) Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiological Reviews 70(2): 567-590.
- France J, Dijkstra J (2005) Volatile fatty acid production. CAB international. pp. 157.
- Nhan NTH, Hon NV, Ngu MT, Von NT, Preston TR, et al. (2001) Practical application of defaunation of cattle on farms in Vietnam: Response of young cattle fed rice straw and grass to a single drench of ground nut oil. Asian-Australian Journnal of Animal Sciences 14(4): 485-490.
- Newbold CJ, Wallace RJ, McIntosh FM (1996) Mode of action of the yeast, Saccharomyces cervisiae as a feed additive for ruminants. Br J Journal of Nutr 76(2): 249-261.
- Hristova AN, Ivan M, Rode LM, McAllister TA (2001): Fermentation characteristics and rumen ciliate protozoal populations in cattle fed medium or high barley-based diet. J Anim Sci 79(2): 515-524.
- Santra A, Karim SA (2002) Influence of ciliate protozoa on biochemical changes and hydrolytic enzyme profile in the rumen ecosystem. J Appl Microbiol 92(5): 801-811
- Jouany JP, Demeyer DI, Grain J (1988) Effect of defaunating the rumen. Animal Feed Science and Technology 21(2-4): 229-265.
- Rasmussen MA, Carlson SA, Franklin SK, McCuddin ZP, Wu MT, et al. (2005) Exposure to rumen protozoa leads to enhancement of pathogenicity of and invasion by multiple-antibiotic-resistant Salmonella enterica bearing SGI1. Infect Immun 73(8): 4668-4675.
- Michalowski T, Rybicka K, Wereszka K, Kasperowicz A (2001) Ability of the Rumen Ciliate Epidinium ecaudatum to Digest and Use Crystalline Cellulose and Xylan for in vitro Growth. ACTA protozoologica 40: 203- 210.
- Singh KM, Ahir VB, Tripathi AK, Ramani UV, Sajnani M, et al. (2012) Metagenomic analysis of Surti buffalo (Bubalus bubalis) rumen: a preliminary study. Mol Biol Rep 39(4): 4841-4848.
- Findley SD, Mormile MR, Sommer-Hurley A, Zhang XC, Tipton P, et al. (2011) Activity-based metagenomic screening and biochemical characterization of bovine ruminal protozoan glycoside hydrolases. Appl Environ Microbiol 77(22): 8106-8113.
- Coleman GS (1986) The metabolism of rumen ciliate protozoa. FEMS Microbiology 39(4): 321-344.
- Orpin CG (1984) The role of ciliate protozoa and fungi in the rumen digestion of plant cell wails. Animal Feed Science and Technology 10(2-3): 121-143.
- Coleman GS (1978) The Metabolism of Cellulose, Glucose and Starch by the Rumen Ciliate Protozoon Eudiplodinium maggii. Journal of General Microbiology 107: 359-366.
- Gijzen HJ, Lubberding HJ, Gerhardus MJT, Vogels GD (1988) Contribution of rumen protozoa to fibre degradation and cellulase activity in vitro. FEMS Microbiology Ecology 53(1): 35-44.
- Bera-Maillet C, Devillard E, Cezette M, Jouany JP, Forano E (2005) Xylanases and carboxymethylcellulases of the rumen protozoa Polyplastron multivesiculatum, Eudiplodinium maggii and Entodinium sp. FEMS Microbiol Lett 244(1): 149-156.
- Devillard E, Newbold CJ, Scott KP, Forano E, Wallace RJ, et al. (1999) A xylanase produced by the rumen anaerobic protozoan Polyplastron multivesiculatum shows close sequence similarity to family 11 xylanase from Gram-positive bacteria. FEMS Microbiol Lett 181(1): 145-152.
- Sebastian R, Kim J, Kim T (2013) Metagenomics: A Promising Approach to Assess Enzymes Biocatalyst for Biofuel Production. Asian Journal of Biotechnology 5(2): 33-50.
- Ferrer M, Golyshina OV, Chernikova TN, Khachane AN, Reyes-Duarte D, et al (2005) Novel hydrolase diversity retrieved from a metagenome library of bovine rumen microflora. Environ Microbiol 7(12): 1996- 2010.
- Uchiyama T, Miyazaki K (2009) Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr Opin Biotechnol 20(6): 616-622.
- Bailey RW (1967) Quantitative studies of ruminant digestion. New Zealand Journal of Agricultural Research 10(1): 15-32.
- Bailey RW, Howard BH (1963) The biochemistry of rumen protozoa. 6. The maltases of Dasytricha ruminantium, Epidinium ecaudatum (Crawley) and Entodinium caudatum. Biochem J 86(3): 446-452.
- Coleman GS (1969) The Metabolism of Starch, Maltose, Glucose and Some Other Sugars by the Rumen Ciliate Entodinium caudatum. J Gen Microbiol 57(3): 303-332.
- Wereszka K, Michalowski T (2012) The ability of the rumen ciliate protozoan Diploplastron affine to digest and ferment starch. Folia microbiologica 57(4): 375-377.
- Bełżecki G, Newbold CJ, McEwan NR, McIntosh FM, Michałowski T (2007) Characterization of the amylolytic properties of the rumen ciliate protozoan Eudiplodinium maggii. Journal of Animal and Feed Sciences 16: 590-606.






























