Adhesiomics - Testing Protein-Protein Interaction of Cell Adhesion Proteins on Microarrays
Qing Yue1, Svenja Bothe1, Sona Mohammadi1, Athanasia Warnecke2, Thomas Scheper3 and Carsten Zeilinger1*
1Department of Biomolecular Drug Research,Leibniz Universität Hannover,Germany
2Department of Otorhinolaryngology,Hannover Medical School,Germany
3Department of Chemistry,Leibniz Universität,Germany
Submission: April 24, 2018; Published: August 27, 2018
*Corresponding author: Carsten Zeilinger,Department ofBiomolecular Drug Research,Institute of Biophysics and Center of Biomolecular Drug Research (BMWZ),Leibniz Universität Hannover, Schneiderberg 38, 30167 Hannover Germany, Email: zeilinger@biophysik.uni-hannover.de
How to cite this article: Qing Yue, Svenja Bothe, Sona Mohammadi, Athanasia Warnecke, Thomas Scheper, Carsten Zeilinger. Adhesiomics - Testing Protein- Protein Interaction of Cell Adhesion Proteins on Microarrays. Adv Biotech & Micro. 2018; 10(3): 555790. DOI: 10.19080/AIBM.2018.10.555790
Keywords
Keywords: Protein; Microarray technology; Synthesized; Heat-shock Proteins; Pathogenic organism; Multicellular organisms; Transmembrane; Immunoglobulin; Epithelia cells; Ovarian; Prognostic marker; Inflammation tissues; Neuronal migration; Biotechnological application; Ambiguous; E.Coli
Abbreviations: HSP:Heat-Shock Proteins; CAM: Cell Adhesion Molecules
Introduction
Protein microarray technology was used recently for target-oriented screening of chemically synthesized compound libraries or isolated natural compounds on Heat-Shock Proteins (Hsp), since Hsps together with the accompanied stress proteome are relevant targets in cancer and pathogenic organism [1-9]. An extended application of this method showed recently that the microarray technology enables also studies of protein-protein interaction to enable a broader application range [4,7]. The basic prerequisite for the formation of multicellular organisms is the ability of a single cell to communicate with neighboring cells. Molecules responsible for this intercellular communication are adhesion molecules like IgCAMs or gap junctions. The adhesion molecules represent a member of the immunoglobulin superfamily of Cell Adhesion Molecules (Ig-CAM). The protein has an extracellular domain, a transmembrane domain, as well as a highly conserved cytoplasmic tail.
L1CAM as a biomarker
L1CAM is a transmembrane cell adhesion molecule belonging to the immunoglobulin superfamily. It has an extracellular six immunoglobulin domains, a transmembrane domain with five type III repeats and cytoplasmic tail [10]. Through its extracellular portion, L1CAM can interact with other L1CAM molecules or other cell adhesion molecules, as well as other different partners, including integrins, proteoglycan, neurocan and CD24 [11]. The expression of L1CAM not only locates in the nervous system, but also indeed in some specialized epithelia such as intestinal crypts, basal and intermediate layers of epidermis and in the epithelia cells of renal and ovarian[12-14].Intensive studies suggest L1CAM is a part of and mediates cellular different signaling pathways and overexpression in various human cancer cells. In the past reports, it has shown that L1CAM expression correlates to poor prognosis, tumor progression and metastasis. For instance, the study of Akts.presented L1CAM is a useful prognostic biomarker and associated with therapeutic effect in primary ovarian cancer. Another study reported the expression of L1CAM is a good prognostic marker of oncogenesis and progression of breast cancer.
Numerous studies exploring the relationship between L1CAM and various carcinoma in vitro and vivo are reported in last three decades. In addition, overexpression of L1CAM was observed in premalignant lesions and inflammation tissues in some reports, that may implicate L1CAM is an early biomarker in the infantile stage of cancer even in the oncogenesis microenvironment before tumor initiation. In a word, L1CAM is associated with development and progression of many types of cancer and is a helpful and promising indicator for diagnosis and treatment of cancer. The understanding of the molecular adhesion process between two cells and corresponding signaling has major impact for medicinal biotechnological application in implant technology, cancer and biofilm research. The neuronal adhesion molecule L1CAM is involved in the development of the nervous system and affects many neuronal processes, such as the growth of axons, neuronal migration and synaptic plasticity[15-17].
The analysis of protein-protein interaction can give insights into the molecular adhesion process with the aim to identify factors influencing the adhesion process of implantation, cancer or biofilm.Since the Hsp network influence a relevant step in the protein maturation protein-protein interaction was analyzed between components of the heat shock network (e.g. Hsp90, Hsp70, PP5, Hsf1) and adhesion protein (L1CAM) or with the adhesion protein itself to visualize weak transient as well strong interaction on the protein microarray. This can help to initiate screenings to study molecular interacting components. The previous elaborate work of other scientists and physician have provided us some useful molecular marker for the diagnosis and therapy of cancer, but the dedicate interaction mechanism of these molecules with other proteins are still ambiguous and a facilitated tool in clinic is required for the easy diagnosis of cancer. Herein, the interactions between promising tumor marker were demonstrated based on microarray and explore the possible secrets of biochemical properties of these markers and the possibility of our microarray as a diagnostic tool.
Protein microarray technology a tool to study proteinprotein interaction
Human proteins of the Hsp90 network and L1CAM have been recombinantly synthesized in E. coli and purified by IMAC. The plasmids containing the target genes of interest were transformed into competent E.coli cells BL21 (DE3). The plasmid of pET15.1 for human Hsp90a [3] and pETSUMO was utilized for Hsp70 [7],Hsf1and L1CAM (commercial) while the plasmids pEXP5-CT containing human TP53(commercial) or human PP5, respectively. Afterwards, E. coli cells were cultivated and expressed the protein. When the cell growth reached the proper OD value (optical density), cell harvest and lysis were performed. The proteins were purified via IMAC and SEC purification and stored at -80℃.All of the proteins were detected on SDS page to check the molecular weight. The proteins were labeled by the dye Cy5 and diluted into required concentration before experiment practice[18,19].
Testing protein-protein interaction on microarrays
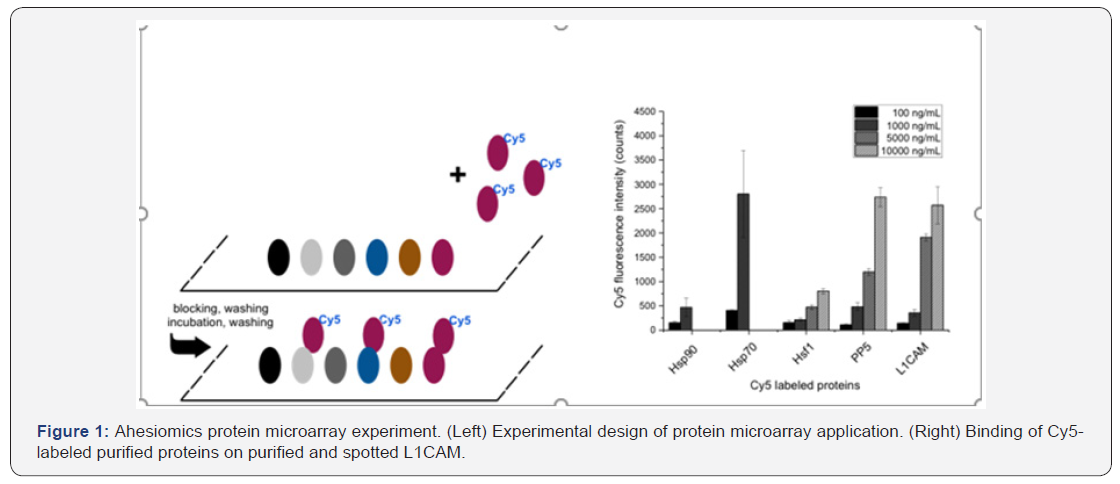
The interaction and binding properties of different proteins or markers were explored based on microarrays approach. First, the proteins and markers were spotted and immobilized on microarrays in a comparable protein concentration (Figure 1, Left). For this purpose, the proteins Hsp90, Hsp70, Hsf1, TP53, PP5, and L1CAM were adjusted to a concentration of 3 mg/ml. Subsequently, the microarrays were blocked by the buffer with 1% BSA to prevent non-specific binding. After three times washing, the microarrays were fixed in a frame and different partners were pipetted into the chamber as the experiment arrangement. Scan and data evaluation proceeded as describedbefore. (Figure 1) (Right) shows dose-responsive binding of Cy5 labeled proteins on spotted L1CAM. Interestingly, Cy5 labeled Hsp70 binds with higher affinity than Cy5 labeled L1CAM and minor activity was found for Hsp90, whereas a weaker affinity was found for PP5 and weak or no affinity for Hsf1.
Discussion and Conclusion
Our data reveal that L1CAM is an interesting target to study different metabolic pathways which can interact with L1CAM. We can show that L1CAM is a candidate for Hsp70 interaction and unknown yet is whether L1CAM plays also a role under cellular stresses like cancer. Several cancer types have elevated Hsp90 and Hsp70 protein levels and act as marker and target for several drugs. It could be that the folding process of L1CAM requires Hsp70 and to hinder protein aggregation due to hydrophobic parts of the protein. This is the first time to our knowledge that the adhesion has been measured on a protein microarray and may be later a tool relevant tool to unravel whether a combinatorial adhesion code in cells exist. Furthermore, it will allow in future compound screening on IgCAMs, interacting bacterial surface factors like CsgA (Curli) and other relevant interacting targets..
Acknowledgement
We are grateful to Prof. Matthias Meyer for plasmids containing human Hsf1 and Prof. Mehdi Mollapur for human PP5.
References
- Kirschning A, Walter J G, Stahl F, Schax E, Scheper T, et al. (2015) Molecular Survival Strategiesof Organisms: HSP and Small Moleculesfor Diagnosticsand Drug Development. Heat Shock Proteins- BasedTherapiesHeat Shock Proteins Book series, Springer Publishers 8, 323-344.
- Franke J, Eichner S, Zeilinger C, Kirschning A (2013) Targeting heatshock- protein 90 (Hsp90) by natural products: geldanamycin, a show case in cancer therapy. Nat Prod Rep 30(10): 1299-1323.
- Schax E, Walter JG, Märzhäuser H, Stahl F, Scheper T, et al. (2014) Microarray-based screening of heats hock protein inhibitors. J Biotechnol 180: 1-9.
- Schax E, Neunaber J, Stahl F, Walter J-G, Scheper T, etal. (2014) Multiplexed heat shock protein micro array as a screening platform for the selection of novel drug compounds. Biodiscovery 14: 1-6.
- Hermane J1, Bułyszko I, Eichner S, Sasse F, Collisi W, et al. (2015) New, non-quin onefluorogeldanamycin derivatives strongly inhibit Hsp 90. Chembiochem. 16(2): 302-311.
- Sharma R, Mohammadi-Ostad-Kalayeh S, Stahl F, Zeilinger C, Dräger G, et al. (2017) Two new labd anediterpenoidsandone new β-lactam from the aerial parts of Royleacinerea. Phytochem. Lett. 19: 101-107.
- Mohammadi-Ostad-Kalayeh S, Hrupins V, Helmsen S, Ahlbrecht C, Stahl F,et al. (2017) Development of a microarray-basedassayforefficientte stingofnew HSP70/DnaKinhibitors. Biorg. Med Chem 25(24): 6345- 6352.
- Mohammadi-Ostad-Kalayeh S, Stahl F, Scheper T, Kock K, Herrmann C, et al. (2018) Heat shock proteins revisit edusing a mutasynthetically generated reblastatinlibrary for comparing the inhibition of human with Leishmania Hsp90s. ChemBioChem. 19:1-14.
- Yue Q, Stahl F, Plettenburg O, Kirschning A, Warnecke A, et al.(2018) Non-competitive effect of gambogicacid displaces fluorescence labelled ATP but requires ATP for binding to Hsp90/HtpG. Biochemistry 57(18):2601-2605.
- Kiefel H, Bondong S, Hazin J, Ridinger J, Schirmer U (2012) L1CAM: a major driver for tumor cell invasion and motility. Cell Adh Migr 6(4): 374-384.
- Angiolini F, Cavallaro U (2007) The pleiotropic role of L1CAM in tumor vasculature. Int J Mol Sci 18 (2): E254.
- Nolte C, Moos M, Schachner M (1999) Immunolocalization of the neural cell adhesion molecule L1 in epithelia of rodents. Cell Tissue Res 298(2): 261-273.
- Debiec H, Christensen EI, Ronco PM (1998) The cell adhesion molecule L1 is developmentally regulated in the renal epithelium and is involved in kidney branching morphogenesis. J Cell Biol 143(7): 2067-2079.
- Zecchini S, Bianchi M, Colombo N, Fasani R, Goisis G, et al. (2008) The differential role of L1 in ovarian carcinoma and normal ovarian surface epithelium. Cancer Res 68(4): 1110-1118.
- Aktas B, Kasimir-Bauer S, Wimberger P, Kimmig R, Heubner M (2013) Utility of mesothelin, L1CAM and afamin as biomarkers in primary ovarian cancer. Anticancer Res 33 (1): 329-336.
- SS Park, SK Park, JH Lim (2011) Expression and prognostic value of L1-CAM in breast cancer. Oncol Rep 25: 223-230.
- Bergmann F, Wands chneider F, Sipos B, Moldenhauer G, Schnie wind B, et al. (2010) Elevated L1CAM expression in precursor lesions and primary and meta stastic tissues of pancreatic ductal adenocarcinoma. Oncol Rep 24(4): 909-915.
- Hentze N, Le Breton L, Wiesner J, Kempf G, Mayer MP (2016) Molecular mechanism of thermosensory function of human heat shock transcription factor Hsf1. Elife 19: 5 pii: e11576.
- Oberoi J, Dunn DM, Wood ford MR, Mariotti L, Schulman J (2016) Structural and functional basis of protein phosphatase 5 substrate specificity. Proc Natl Acad Sci U S A. 113(32): 9009-9014.






























