Enhanced Coenzyme Q10 Yield by Blocking Hopanoids Pathway
Wen Xu, Jia Yao, Xi Ma, Lingqiao Shao and Yang Wang*
Department of Pathogen Biology, Xi'an Medical University, China
Submission: March 03, 2017; Published: April 26, 2017
*Corresponding author: Yang Wang, Department of Pathogen Biology, School of Medical Science, Xi'an Medical University, Xi'an 710021, Shaanxi, China, Tel: 86-29-86131371; Email: yang.wang@xiyi.edu.cn
How to cite this article: Wen Xu, J Y, Xi Ma, Lingqiao S Yang W. Enhanced Coenzyme Q10 Yield by Blocking Hopanoids Pathway. Adv Biotech & Micro. 2017; 3(2): 555610.DOI: 10.19080/AIBM.2017.03.555610
Abstract
Microbial synthesis of coenzyme Q10 (CoQ10) by fermentation processes has been emerging in recent years. Given that the synthesis of hopanoids occurs as a branched pathway of CoQ10 synthesis, we hypothesized that blocking the hopanoids pathway might improve the CoQ10 yield. The recombinant strain R. palustris (Ashc) with blocked hopanoids pathway was employed for studying the accumulation of CoQ10. It was found that the CoQ10 content in R. palustris (Ashc) reached 3.71mg/g DCW, corresponding to 34.7% improvement over the wild type strain.Microbial synthesis of coenzyme Q10 (CoQ10) by fermentation processes has been emerging in recent years. Given that the synthesis of hopanoids occurs as a branched pathway of CoQ10 synthesis, we hypothesized that blocking the hopanoids pathway might improve the CoQ10 yield. The recombinant strain R. palustris (Ashc) with blocked hopanoids pathway was employed for studying the accumulation of CoQ10. It was found that the CoQ10 content in R. palustris (Ashc) reached 3.71mg/g DCW, corresponding to 34.7% improvement over the wild type strain.
Moreover, rate-limiting enzymes including endogenous UbiA and Dps from Rhodobacter sphaeroides were co-expressed, and the resulting strain yielded an additional 94% improvement of CoQ10 content over strain R. palustris (Ashc). Upon supplementing with NaHSO3, the CoQ10 content was further increased to 6.51 mg/g DCW and the biomass was enhanced from 1.07g/l to 1.33g/l. The effectively strategies represented here will enhance our ability to design the system with metabolic engineering techniques for higher CoQ10 production.
Keywords: Coenzyme Q10; Co-expression; Hopanoids; Pathway blocking; Rhodopseudomonas palustris
Abbreviations: CoQ10: Coenzyme Q10; DCW: Dry Cell weight; DPP: Decaprenyl Diphosphate; FPP: Farnesyl Diphosphate; Phba: Parahydroxybenzoic Acid; UQ: Ubiquinone; PNSB: Purple Non-Sulfur Bacteria
Introduction
Coenzyme Q10 (CoQ10), otherwise known as ubiquinone (UQ), is a valuable bioactive compound used both medically and cosmetically [1]. Many studies focus on enhancing CoQ10 production in Escherichia coli (E.coli) by genetic engineering [2-9]. Agrobacterium tumefaciens also have been employed as the producers of CoQ10 [10,11]. Typical purple non-sulfur bacteria (PNSB) generally have a relatively high content of CoQ10, as ideal CoQ10 producers [12-16]. R. Palustris (Rhodopseudomonas palustris) is a PNSB with a high content of inherent CoQ10, whose fermentation processes have been well researched over the years [17,18]. Therefore, R. Palustris could be a potential host for CoQ10 production by metabolic engineering.Coenzyme Q10 (CoQ10), otherwise known as ubiquinone (UQ), is a valuable bioactive compound used both medically and cosmetically [1]. Many studies focus on enhancing CoQ10 production in Escherichia coli (E.coli) by genetic engineering [2-9]. Agrobacterium tumefaciens also have been employed as the producers of CoQ10 [10,11]. Typical purple non-sulfur bacteria (PNSB) generally have a relatively high content of CoQ10, as ideal CoQ10 producers [12-16]. R. Palustris (Rhodopseudomonas palustris) is a PNSB with a high content of inherent CoQ10, whose fermentation processes have been well researched over the years [17,18]. Therefore, R. Palustris could be a potential host for CoQ10 production by metabolic engineering.
R. palustris TIE-1 contains more than 30mg/l DCW (dry cell weight) of hopanoids that are not required for growth under normal conditions, although they play a role in membrane integrity and pH homeostasis [19]. The deletion of squalene- hopene cyclase protein (Shc), which cyclizes squalene to the basic hopene structure, can make the strain no longer produce any hopanoids [19]. As the synthesis of hopanoids occurs as a branched pathway of CoQ10 synthesis in R. palustris TIE- 1 (Figure 1), it was hypothesized that blocking the hopanoids pathway might direct FPP flux towards CoQ10 pathway.R. palustris TIE-1 contains more than 30mg/l DCW (dry cell weight) of hopanoids that are not required for growth under normal conditions, although they play a role in membrane integrity and pH homeostasis [19]. The deletion of squalene- hopene cyclase protein (Shc), which cyclizes squalene to the basic hopene structure, can make the strain no longer produce any hopanoids [19]. As the synthesis of hopanoids occurs as a branched pathway of CoQ10 synthesis in R. palustris TIE- 1 (Figure 1), it was hypothesized that blocking the hopanoids pathway might direct FPP flux towards CoQ10 pathway.
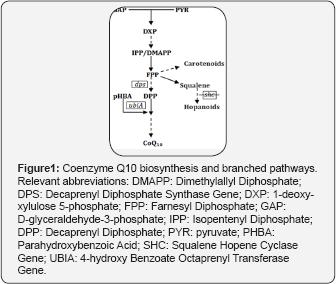
Materials and Methods
Medium, culture conditions and general methods
The bacteria seed cultivation was performed at 30 0c in YP medium (3g/l peptone, 3/l yeast extract, pH 6.5) for 48h under aerobic condition. The engineered R. palustris strains for squalene overproduction were cultured at 30 0c in a designed medium (2g/l sodium succinate, 10g/l glucose, 3g/l peptone, 3g/l yeast extract, 2g/l KH2PO4, 2g/l K2HPO4, 2g/l MgSO•7H2O, 0.5g/l FeSO4.7H2O, 5ml/l mineral solution, pH 6.5) under anaerobic condition. The mineral solution contained: 1g/l NaMoO.2H2O, 2g/l CuSO.5H2O, 1g/l ZnSO4, 1g/l H3BO3, 1g/l MnCl2. Anaerobic cultivation was achieved via static culture in the presence of nitrogen with white fluorescent light (3200 lx). Kanamycin of 350mg/l was supplemented to the culture media to retain the constructed plasmids with corresponding antibiotic selection markers. All the cell cultures were grown for 120h to be in the stationary phase, before the cells were collected for CoQ10 analysis.
General molecular manipulations were performed according to standard protocols. PCRs were performed using PrimeSTAR HS DNA polymerase (TaKaRa, Dalian City, China). Electroporator (Eppendorf, Humburg, Germany) was utilized for transforming the constructed plasmids into the R. palustris strains (12.5kv/ cm, 200 Ω, 25 μF).
plasmid construction
For gene overexpression, the primers F_dps_EcoRI (GGAATTCGTGAATGGGATTGGACGAG GTTTCG) and R_dps_ BamHI (CGGGATCCTCAGGCGATGCGTTCGACCA) were used to amplify dps gene from Rhodobacter sphaeroides. And then, the gene segment was ligated into pMG103 between EcoR I and BamH I sites to generate the plasmid pMGD. The ubiA gene was amplified with the primers F_ubiA_BamHI (CGGGATCCATGAGTGGAATTCCGGCCAG) and R_ubiA_ HindIII (CCCAAGC TTTCACGCCATGCTGCGCGAGA) containing BamHI and HindIII sites and ligated into pMGD to generate pMGDU.
coQ10 and lycopene assay
Cell growth was measured using a spectrophotometer at 600nm and converted to dry cell weight using a prepared standard curve of DCW versus 0D600.Vol. CoQ10 was extracted with an organic solvent from the saponified liquid before being subjected to high-performance liquid chromatography (Hitachi, Tokyo, Japan), according to the procedure described before [13]. The extraction of lycopene was carried out according to previous research [24], its content was estimated via HPLC at 475nm [25].
Results and Discussion
increased CoQ10 content in R. palustris (Δshc)
For accumulating squalene in R. palustris, the shc gene was deleted to block hopanoids pathway in our previous study [26]. Although the titer of squalene in R. palustris (Δshc) reached 3.8mg/g DCW, it was much lower than the yield of total hopanoids (about 30mg/g DCW) in the wild strain, suggesting that many FPPs had been directed into other pathways. Here, R. palustris (Δshc) was employed to study the accumulation of CoQ10 and carotenoids. Lycopene, a metabolite in the carotenoids pathway, was analyzed to represent the accumulation of carotenoids.
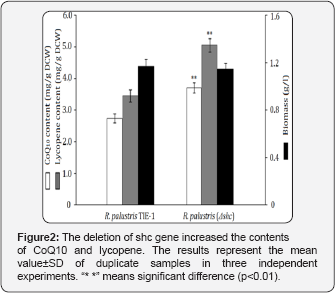
As shown in Figure 2, the CoQ10 content of R. palustris (Δshc) reached 3.71mg/g DCW, which corresponds to a 34.7% improvement over the parental strain R. palustris TIE-1. Moreover, R. palustris (Δshc) yielded a 46.2% improvement of the lycopene content over R. palustris TIE-1. These results suggested that shc deletion diverted the FPP flux from the production of hopanoids to the biosyntheses of both CoQ10 and carotenoids. It is noteworthy that there was not a noticeable decrease in biomass of the recombinant strain, demonstrating the hopanoids pathway blocking was an efficient strategy for improving CoQ10 production. However, carotenoids content increased more significantly, means that the increase in CoQ10 production might be restricted by the increased carotenoids yield because it consumed more FPP (Figure 2).
CoQ10 production improvement by the co-expression of dps and ubiA
To divert more FPP to the biosynthesis of CoQ10, decaprenyl diphosphate synthase (Dps) from Rhodobacter sphaeroides 2.4.1 which catalyzes FPP to form DPP was expressed. The investigation revealed that the CoQ10 content of R. palustris (Δshc)/pMGD was increased to 4.38mg/g DCW but the CoQ10 content in R. palustris TIE-1/pMGD only reached 3.15mg/g DCW (Table 1). It was clear that the increment of CoQ10 content in R. palustris (Δshc)/pMGD was higher than that in R. palustris TIE- 1/pMGD. In the meantime the lycopene content in R. palustris (Δshc)/pMGD was decreased from 5.06mg/g DCW to 4.67mg/g DCW Table 1). Results suggested that the Dps expression enhanced FPP flux to the biosynthesis of CoQ10 and decreased the FPP flux to carotenoids pathway. However, the increment of CoQ10 content was not as high as expected. It was hypothesized that the limited pools of pHBA and UbiA restricted the utilization of DPP for CoQ10 biosynthesis, although more DPP had been formed by Dps expression.
UbiA which catalyzes pHBA and DPP to form decaprenyl- pHBA is one of the rate-limiting enzymes in the synthetic pathway of CoQ10. And its overexpression has been shown to increase CoQ10 content, especially when accompanied with the supplementation of pHBA [2,6]. To increase the utilization of DPP for CoQ10 biosynthesis, endogenous UbiA was over expressed along with the pHBA supplementation (100mg/l). As can be seen in Table 1, the CoQ10 content was increased to 6.16mg/g DCW in R. palustris (Δshc)/pMGDU, while that was 4.42mg/g DCW for R. palustris TIE-1/pMGDU. Notably, the lycopene content was further decreased to 3.92 mg/g DCW. Obviously, the expression of UbiA and supplementation with pHBA enhanced the competitiveness of CoQ10 pathway for FPP. However, the cell mass was slightly decreased, which may caused by metabolic burden due to protein overexpression (Table 1).

WT, R. palustris TIE-1; Dshc, R. palustris (Δshc); CK, control; +, expression or supplementation; Data represent the mean value±SD of duplicate samples from three separate experiments.
NaHSO3 enhanced both CoQ10 content and biomass
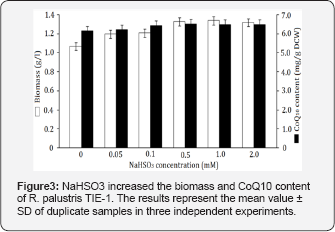
To study the influence of NaHSO3 on the cell growth and CoQ10 production of R. palustris TIE-1, NaHSO3 at different levels was supplemented into the culture medium. As shown in Figure 3, both the biomass and the CoQ10 content were increased in the presence of NaHSO3. The supplementation with 0.5mM NaHSO3 resulted in 6.51mg/g DCW CoQ10 and 1.33g/l biomass, generating the highest CoQ10 production of 8.65mg/g, corresponding to a 33.2% improvement over the control (Figure 3). The increases in biomass and CoQ10 content indicated that NaHSO3 increased the photo-phosphorylation with intensified electron transport efficiency, and thus the accumulation of CoQ10 was enhanced as one of the electron transfer agents. In future, more studies are needed to explore the functional mechanism of NaHSO3 (Figure 3).
Conclusion
In this study, we report several strategies, including hopanoids pathway blocking, genes co-expression and NaHSO3 supplementation, for enhancing CoQ10 production. Based on the combination of these strategies, the content of CoQ10 reached 6.51mg/g DCW, which was 1.3-times higher than that for wildtype strain. This work enriched the strategy for metabolic engineering and showed the potential of producing coenzyme Q10 by Rhodopseudomonas palustris TIE-1.In this study, we report several strategies, including hopanoids pathway blocking, genes co-expression and NaHSO3 supplementation, for enhancing CoQ10 production. Based on the combination of these strategies, the content of CoQ10 reached 6.51mg/g DCW, which was 1.3-times higher than that for wildtype strain. This work enriched the strategy for metabolic engineering and showed the potential of producing coenzyme Q10 by Rhodopseudomonas palustris TIE-1.
Acknowledgment
The authors would like to gratefully acknowledge Dianne K. Newman for providing the Rhodopseudomonas palustris TIE-1 strain and Masayuki Inui for providing the plasmid pMG103. This paper was supported by the Shaanxi Science and Technology Innovation Project 2016KTCQ03-07 (to Y. Wang).The authors would like to gratefully acknowledge Dianne K. Newman for providing the Rhodopseudomonas palustris TIE-1 strain and Masayuki Inui for providing the plasmid pMG103. This paper was supported by the Shaanxi Science and Technology Innovation Project 2016KTCQ03-07 (to Y. Wang).
References
- Sharma S, Kheradpezhou M, Shavali S, El Refaey H, Eken J, et al. (2004) Neuroprotective actions of coenzyme Q10 in Parkinson's disease. Quinones and Quinone Enzymes 382: 488-509.
- Kim SJ, Kim MD, Choi JH, Kim SY, Ryu YW, et al. (2006) Amplification of 1-deoxy-D-xyluose 5-phosphate (DXP) synthase level increases coenzyme Q10 production in recombinant Escherichia coli. Appl Microbiol Biotechnol 72(5): 982-985.
- Zahiri HS, Yoon SH, Keasling JD, Lee SH, Kim SW, et al. (2006) Coenzyme Q10 production in recombinant Escherichia coli strains engineered with a heterologous decaprenyl diphosphate synthase gene and foreign mevalonate pathway. Metab Eng 8(5): 406-416.
- Seo MJ, Im EM, Nam JY, Kim SO (2007) Increase of CoQ10 production level by the coexpression of decaprenyl diphosphate synthase and 1-Deoxy-D-xylulose 5-phosphate synthase isolated from Rhizobium radiobacter ATCC 4718 in recombinant Escherichia coli. J Microbiol Biotechnol 17(6): 1045-1048.
- Cluis CP, Burja AM, Martin VJJ (2007) Current prospects for the production of coenzyme Q10 in microbes. Trends Biotechnol 25(11): 514-521.
- Cluis CP, Ekins A, Narcross L, Jiang H, Gold ND, et al. (2011) Identification of bottlenecks in Escherichia coli engineered for the production of CoQ10. Metab Eng 13(6): 733-744.
- Park YC, Kim SJ, Choi JH, Lee WH, Park KM, et al. (2005) Batch and fed-batch production of coenzyme Q10 in recombinant Escherichia coli containing the decaprenyl diphosphate synthase gene from Gluconobacter suboxydans. Appl Microbiol Biotechnol 67(2): 192-196.
- Martinez I, Méndez C, Berrios J, Altamirano C, Diaz-Barrera A (2015) Batch production of coenzyme Q10 by recombinant Escherichia coli containing the decaprenyl diphosphate synthase gene from Sphingomonas baekryungensis. J Ind Microbiol Biotechnol 42(9): 1283-1289.
- Xu W, Yang S, Zhao J, Su T, Zhao L, et al. (2014) Improving coenzyme Q8 production in Escherichia coli employing multiple strategies. J Ind Microbiol Biotechnol 41(8): 1297-1303.
- Ha SJ, Kim SY, Seo JH, Moon HJ, Lee KM, et al. (2007) Controlling the sucrose concentration increases Coenzyme Q10 production in fed-batch culture of Agrobacterium tumefaciens. Appl Microbiol Biotechnol 76(1): 109-116.
- Gu SB, Yao JM, Yuan QP, Xue PJ, Zheng ZM, et al. (2006) Kinetics of Agrobacterium tumefaciens ubiquinone-10 batch production. Process Biochem 41(8): 1908-1912.
- Yoshida H, Kotani Y, Ochiai K, Araki K (1998) Production of ubiquinone-10 using bacteria. J Gen Appl Microbiol 44(1): 19-26.
- Kien NB, Kong IS, Lee MG, Kim JK (2010) Coenzyme Q10 production in a 150-l reactor by a mutant strain of Rhodobacter sphaeroides. J Ind Microbiol Biotechnol 37(5): 521-529.
- Lu W, Ye L, Xu H, Xie W, Gu J, et al. (2014) Enhanced Production of Coenzyme Q10 by Self-Regulating the Engineered MEP Pathway in Rhodobacter sphaeroides. Biotechnol Bioeng 111(4): 761-769.
- Lu W, Ye L, Lv X, Xie W, Gu J, et al. (2015) Identification and elimination of metabolic bottlenecks in the quinone modification pathway for enhanced coenzyme Q10 production in Rhodobacter sphaeroides. Metabolic Engineering 29: 208-216.
- Tiana Y, Yuea T, Yuana Y, Somab PK, Loa YM (2010) Improvement of cultivation medium for enhanced production of coenzyme Q10 by photosynthetic Rhodospirillum rubrum. Biochem Eng J 51(3): 160166.
- Carlozzi P, Sacchi A (2001) Biomass production and studies on Rhodopseudomonas palustris grown in an outdoor, temperature controlled, underwater tubular photobioreactor. J Biotechnol 88(3): 239-249.
- Carlozzi P, Pushparaj B, Degl'Innocenti A, Capperucci A (2006) Growth characteristics of Rhodopseudomonas palustris cultured outdoors, in an underwater tubular photobioreactor, and investigation on photosynthetic efficiency. Appl Microbiol Biotechnol 73(4): 789-795.
- Welander PV, Hunter RC, Zhang L, Sessions AL, Summons RE, et al. (2009) Hopanoids play a role in membrane integrity and pH homeostasis in Rhodopseudomonas palustris TIE-1. J Bacteriol 191(19): 6145-6156.
- Ji BH, Tan HH, Zhou R, Jiao DM, Shen YG (2005) Promotive effect of low concentrations of NaHSO3 on photophosphorylation and photosynthesis in phosphoenolpyruvate carboxylase transgenic rice leaves. Journal of Integrative Plant Biology 47(2): 178-186.
- Wu Y, He W, Ma W, Shen Y, Mi H (2012) Low concentrations of NaHSO3 enhance NADPH dehydrogenase-dependent cyclic photophosphorylation and alleviate the oxidative damage to improve photosynthesis in tobacco. Chin Sci Bull 57(30): 3872-3877.
- Wang HW, Mi HL, Ye JY, Deng Y, Shen YK (2003) Low concentrations of NaHSO3 increase cyclic photophosphorylation and photosynthesis in Cyanobacterium Synechocystis PCC6803. Photosynth Res 75(2): 151159.
- Evans MCW (1979) Photosynthetic bacteria. Nature 279(5713): 561.
- Alper H, Jin YS, Moxley JF, Stephanopoulos G (2005) Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab Eng 7(3): 155-164.
- Kim SW, Keasling J (2001) Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol Bioeng 72(4): 408-415.
- Xu W, Yuan J, Yang S, Ching CB, Liu J (2016) Programming Saposin- Mediated Compensatory Metabolic Sinks for Enhanced Ubiquinone Production. ACS synthetic biology 5(12): 1404-1411.






























