Exploring Variation in Structure and Intrinsic Disorder of Lactoferrampin among Different Animal Species
Nawal Abd El-Baky and Amro A Amara*
Protein Research Department, Genetic Engineering and Biotechnology Research Institute (GEBRI), City of Scientific Research and Technological Applications (SRTA-City), New Borg El-Arab City, Alexandria, Egypt
Submission: December 07, 2023;Published: December 21, 2023
*Corresponding author: Amro A Amara, Protein Research Department, Genetic Engineering and Biotechnology Research Institute (GEBRI), City of Scientific Research and Technological Applications (SRTA-City), New Borg El Arab City, Alexandria, Egypt
How to cite this article: El-Baky N. A., Amara A. A. Exploring Variation in Structure and Intrinsic Disorder of Lactoferrampin among Different Animal Species. Arch Anim Poult Sci. 2023; 2(4): 555591. DOI: 10.19080/AAPS.2023.04.555591
Abstract
Lactoferrampin (Lfampin) is second in order of importance among bioactive peptides derived from lactoferrin (Lf), a protein with multiple functions found chiefly in mammalian milk. This peptide has two shared features with other antimicrobial peptides; a net positive charge, and an amphipathic character. Lfampin linked to lactoferricin (another bioactive peptide derived from Lf) can be an efficient substitute to antibiotics in feed. Determining the variation in structure and intrinsic disorder of Lfampin among different animal species and human is vital to elucidate its antimicrobial activities. In this study, the amino acid sequences of parent protein of this peptide from 60 animal species and human were obtained from NCBI database and analyzed by multiple alignment, and phylogenetic tree of animal species was built. Lfampin was located in this alignment of parent protein of eighteen different animal species besides human, and then aligned individually. The variation in intrinsic disorder and disorder propensity was explored.
The hydrophobicity and charge characteristics of Lfampin primary structure in eighteen animal species and human were also determined. The region that includes the peptide displayed a range of 62.8% (Sus scrofa) to 97.1% (Camelus ferus) identity relative to Camelus dromedarius among 18 animal species and human. The variation in structure of the region that includes Lfampin is also correlated with diverse levels of intrinsic disorder as well as disorder propensity in its sequence among different animal species and human. Interestingly, no intrinsic disorder was detected in camel (Camelus ferus, and Camelus dromedarius) Lfampin or the 35-residues region that contains the peptide. Chlorocebus sabaeus Lfampin has zero net positive charge, while Bos indicus, Bos grunniens, and Bos taurus peptides have the highest net charge (=5). This high net charge can anticipate higher antimicrobial effects. Distribution of intrinsic disorder in bioactive proteins/peptides can identify their source such as animal milk source. Furthermore, intrinsic disorder can predict strength of peptide activity for its application in feed or food and as peptide drug.
Keywords:Predatory bacteria; Aquaculture; Antibiotic substitution; Probiotics; Disease control
Introduction
Lactoferrampin is one of two antimicrobial peptides found in N-terminal region of Lf parent protein and comprises seventeen amino acids [1]. The parent protein of this peptide is involved in vast range of bioactivities from antimicrobial effects that are either based on its capacity to bind iron or not related to this capacity to immune system triggering [2-4]. These parent protein activities are mostly endorsed by its N-terminal region and high cationicity of this region [5].
The N-terminal sequence of Lf comprises two efficient bioactive peptides; lactoferricin (Lfcin) and Lfampin. Lfcin is released via pepsin hydrolysis of the parent protein [6]. Lfcin performs numerous activities comprising antimicrobial and immunological effects [7]. Lfampin is positioned in close proximity to Lfcin. The sequence of bovine Lfampin corresponds directly to residues 268–284 (WKLLSKAQEKFGKNKSR) in parent protein cationic N-terminal domain [1]. The structure of this peptide promotes its potential application as an antimicrobial peptide due to its net cationic charge and the existence of tryptophan in its sequence [8-10]. Bovine Lfampin could act against Gram-negative bacterial strains, Gram-positive bacterial strains, along with Candida albicans [1].
Lfampin linked to Lfcin can be applied as an efficient substitute to antibiotics (e.g., colistin sulfate) in feed of piglets weaned at age of three weeks to enhance their growth performance [11,12]. Both Lfampin and Lfcin possess superior antimicrobial effects than their parent protein [13] could engineer food-grade Lactococcus lactis expressing a chimeric peptide of Lfampin linked to Lfcin that has antimicrobial effects and can be applied as bio-preservative in food industry [14].
Predicting intrinsic disorder in proteins is vital since it performs a wide range of functions in the cell. In spite of absence of stable 3D structure in these disordered proteins, they are responsible for countless functions in cell [15-17]. Research revealed that disordered region’s primary structure varies considerably from that of ordered ones [18]. Thus, several prediction approaches were developed to predict disorder depending on the sequence of amino acids. RONN [19], GlobPlot [20], Predictor of Natural Disordered Regions (PONDR) [21], DISOPRED [22,23], DISEMBL [24], VL3 [25], and IUPred [26] can predict the likelihood that a definite amino acid residue is found in a disordered region via knowing the sequences of amino acids close to that residue. On the other hand, there are other methods that can predict disorder via amino acid sequences binary grouping into typically disordered sequences and typically ordered ones [27-29].
The aim of this work was to determine the variation in structure, hydrophobicity and charge characteristics of primary structure, intrinsic disorder, and disorder propensity of Lfampin among eighteen different animal species and human to elucidate its antimicrobial activities.
Materials and Methods
Protein sequences
The amino acid sequences of parent protein (Lf) of Lfampin from different animal species were searched in NCBI database (NCBI Blast: Protein Sequence, last accessed on 11/28/2023) via BLASTP program. The following sequence of camel parent protein (Accession: AHJ37525) was used as Query sequence in this search.
Camelus dromedarius Lf
Length: 708 amino acids
˂MKLFFPALLSLGALGLCLAASKKSVRWCTTSPAESSKCAQWQRRMKKVRGPSVTCVKKTSRFECIQAISTEKADAVTLDGGLVYDAGLDPYKLRPIAAEVYGTENNPQTHYYAVAIAKKGTNFQLNQLQGLKSCHTGLGRSAGWNIPMGLLRPFLDWTGPPEPLQKAVAKFFSASCVPCVDGKEYPNLCQLCAGTGENKCACSSQEPYFGYSGAFKCLQDGAGDVAFVKDSTVFESLPAKADRDQYELLCPNNTRKPVDAFQECHLARVPSHAVVARSVNGKEDLIWKLLVKAQEKFGRGKPSAFQLFGSPAGQKDLLFKDSALGLLRIPSKIDSGLYLGSNYITAIRGLRETAAEVELRRAQVVWCAVGSDEQLKCQEWSRQSNQSVVCATASTTEDCIALVLKGEADALSLDGGYIYIAGKCGLVPVLAESQQSPESSGLDCVHRPVKGYLAVAVVRKANDKITWNSLRGKKSCHTAVDRTAGWNIPMGLLFKNTDSCRFDEFFSQSCAPGSDPRSKLCALCAGNEEGQNKCVPNSSERYYGYTGAFRCLAENVGDVAFVKDVTVLDNTDGKNTEQWAKDLKLGDFELLCLNGTRKPVTEAESCHLAVAPNHAVVSRIDKVAHLEQVLLRQQAHFGRNGQDCPGKFCLFQSKTKNLLFNDNTECLAKLQGKTTYEEYLGPQYVTAIAKLRRCSTSPLLEACAFLMR˃
Adjustment of protein sequences and their alignment
After NCBI database search was accomplished, the top 113 Blast hits for Lf protein sequences from different mammals were generated. Repeated or similar sequences were removed and only 78 protein sequences were selected. These sequences were saved in FASTA format for additional exploration. Inspecting these 78 sequences, it could be concluded that they are derived from 60 different animal species and human Table 1. The sequences obtained from NCBI database were aligned using Molecular Evolutionary Genetics Analysis Version 11 (MEGA11) [30]. The sequence identity of 78 lactoferrin sequences obtained from NCBI database relative to Camelus dromedarius (Accession: AHJ37525) was calculated. MEGA11 was also applied to build phylogenetic tree of parent protein among different animal species.
Peptide sequences and their alignment
Lfampin was located in the alignment of parent protein of different animal species. The 35-residues region of Lfampin (the sequence of amino acids that corresponds directly to formerly identified Lfampin derived from bovine lactoferrin (17 amino acids) with added 18 residues near C-terminal end of peptide from the parent protein sequence) was aligned individually among 18 animal species and human using BioEdit version 7 [31]. The selected 18 animal species include Camelus dromedarius (Arabian camel, even-toed ungulates), Camelus ferus (wild Bactrian camel, even-toed ungulates), Physeter catodon (Sperm whale, whales & dolphins), Lagenorhynchus albirostris (White-beaked dolphin, whales & dolphins), Equus asinus (Ass, odd-toed ungulates), Phocoena sinus (Vaquita, whales & dolphins), Myotis brandtii (Brandt's bat, bats), Miniopterus natalensis (bats), Bos grunniens (Domestic yak, even-toed ungulates), Bos indicus (Zebu cattle, even-toed ungulates), Trachypithecus francoisi (Francois's langur, primates), Gorilla gorilla gorilla (Western lowland gorilla, primates), Budorcas taxicolor (Takin, even-toed ungulates), Sus scrofa (Pig, even-toed ungulates), Capra hircus (Goat, even-toed ungulates), Chlorocebus sabaeus (Green monkey, primates), Bos taurus (Cattle, Artiodactyla), and Oryx dammah (Scimitar-horned oryx, even-toed ungulates). Sequence identity of 20 sequences of Lfampin region relative to Camelus dromedarius Lfampin region was determined.
Prediction of natural disordered regions and disorder propensity in Lfampin
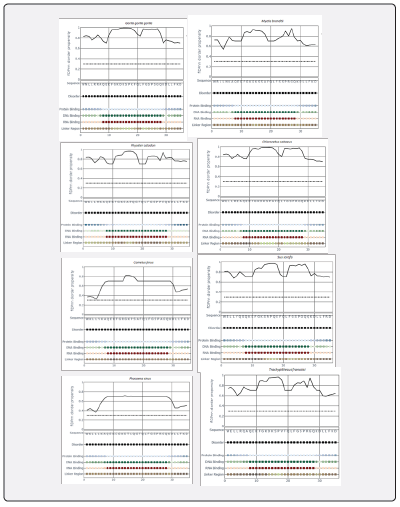
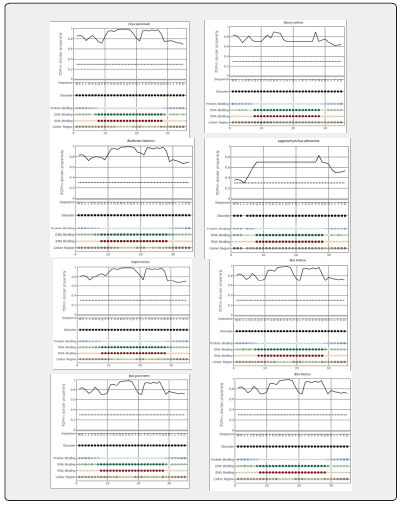
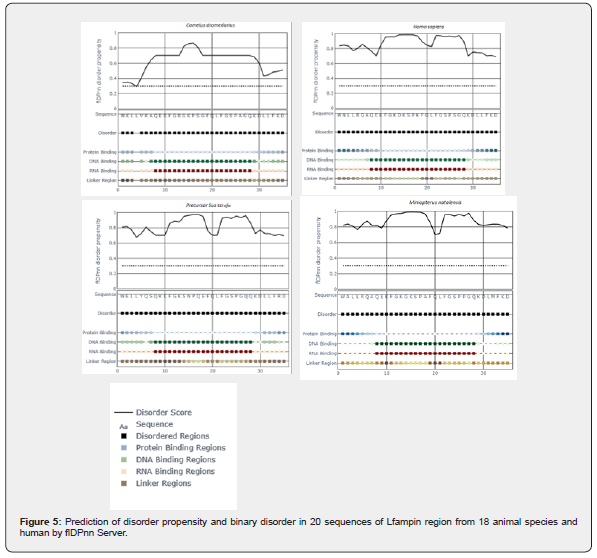
Prediction of natural disordered regions in 20 amino acid sequences of 35-residues region of Lfampin from 18 animal species and human was done by PONDR (www.pondr.com, last accessed on 11/29/2023). Furthermore, prediction of disorder propensity and binary disorder in 20 sequences of Lfampin region from 18 animal species and human was performed via flDPnn Server (biomine.cs.vcu.edu/webresults/flDPnn/20231129035754/results.html, last accessed on 11/29/2023). The flDPnn Server generated the following data: binary disorder (value of 1 represents disordered residue, and 0 represents ordered residue), disorder propensity (greater value signifies higher possibility that a certain residue is disordered), binary protein-binding (value of 1 represents disordered protein-binding residue, 0 represents other disordered residue, and X represents ordered residue), protein-binding propensity, binary DNA-binding, DNA-binding propensity, binary RNA-binding, RNA-binding propensity, binary linker, and linker propensity.
Hydrophobicity and charge characteristics of Lfampin primary structure
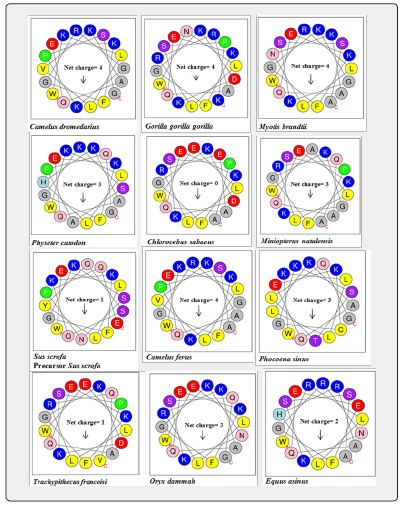
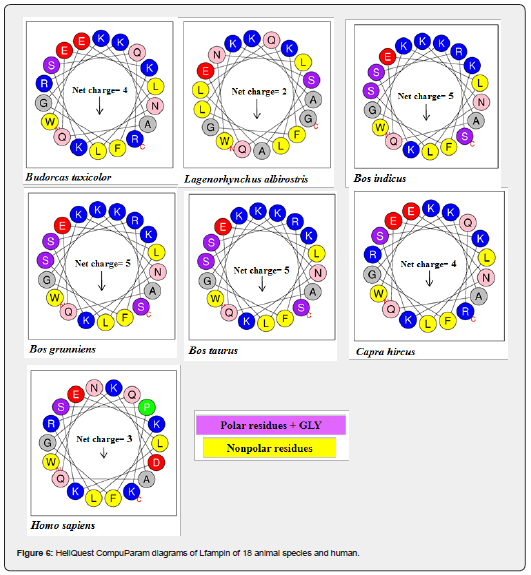
The primary structure of 20 amino acid sequences of Lfampin from 18 animal species and human was analyzed by HeliQuest CompuParam version3 (https://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParams.py, last accessed on 12/2/2023). This analysis generated different characteristics of the peptide including physico-chemical properties (hydrophobicity, hydrophobic moment, and net positive charge), polar and nonpolar residues, uncharged and charged residues, aromatic residues, and special residues.
Results and Discussion
Despite the fact that Lfampin is neighboring Lfcin in cationic N-terminal domain of their parent protein and have comparable cationic and amphipathic features, their ability to kill bacterial pathogens differs from each other. The reason for this difference in their antibacterial activity is the significant variation in their amino acid composition and chain length, and consequently great variation in their structures [5]. Lfampin has a crucial contribution to Lf effects on bacterial membrane [1]. Lfampin can kill various Gram-negative bacterial strains, Gram-positive bacterial strains, parasites, and yeasts as reported in MilkAMP database (a comprehensive database of antimicrobial peptides of dairy origin) [1,32].
The comparison of sequence and primary structure of Lfampin from different animal species and human is important to elucidate the peptide antimicrobial action and predict which one is a candidate for developing therapeutic peptide. The present study provides the first comprehensive description of sequence, hydrophobicity, and charge characteristics of primary structure of Lfampin from 18 different animal species and human.
MEGA11 alignment of 78 sequences obtained from NCBI database of parent protein (Lf) from 60 animal species and human is demonstrated in Figure 1. The amino acid sequences of lactoferrin in 60 different animal species and human showed a range of identity of 73.9% (Homo sapiens) to 99.1% (Camelus ferus, and Camelus bactrianus) relative to lactoferrin of Camelus dromedarius Table 2. Figure 2 illustrates phylogenetic tree built by MEGA11 of parent protein (Lf) among different animal species.
BioEdit alignment of amino acid sequences of the 35-residues Lfampin region from 18 animal species and human is shown in Figure 3. The identity results of the 35-residues region that contains Lfampin differ from those of its parent protein or lactoferrin. The 35-residues region that includes the peptide displayed a range of 62.8% (Sus scrofa) to 97.1% (Camelus ferus) identity relative to Camelus dromedarius among 18 animal species and human Table 3. Data in Figure 3 and Table 3 support the presence of significant variance in sequence of the 35-residues region that includes Lfampin among 18 different animal species and human, which can lead to differences in its structure, hydrophobicity, charge characteristics, intrinsic disorder, disorder propensity, and thus its biological activities.
Disordered proteins or peptides are receiving raising attention since a lot of of them are functionally essential. However, disorder information for a given protein or peptide is mostly extracted from X-ray analysis of its crystal. This means that most proteins that are difficult to crystallize or those not crystallized yet even if they are disordered will not be reported. Therefore, intrinsic disorder prediction from protein sequences that have been experimentally determined based on difference from disorder among structure-unknown protein data will benefit reporting more disordered proteins [17]. PONDR algorithms are among the methods that predict disorder depending on structure-known records from already crystallized proteins [21].
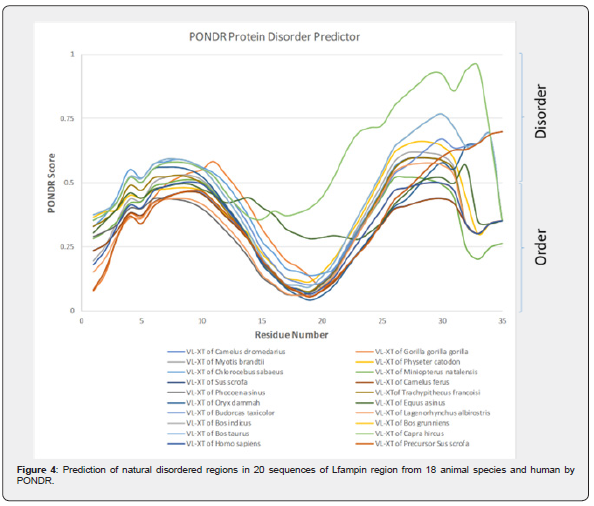
Prediction of natural disordered regions in 20 sequences of Lfampin region from 18 animal species and human performed by PONDR is illustrated in Figure 4 revealed that the highest PONDR score was for Capra hircus (goat Lfampin region), followed by 3 entirely overlapped curves for PONDR score representing Bos grunniens (domestic yak Lfampin region), Bos indicus (Zebu cattle Lfampin region), and Bos taurus (cattle Lfampin region). PONDR VLXT NNP statistics of 20 sequences of Lfampin region in Table 4 demonstrated an interesting and surprising fact: both Camelus dromedarius and Camelus ferus have zero number of disordered regions, zero overall percent of disorder, and that 17-residues Lfampin along with 35-residues Lfampin region are ordered. Physeter catodon, Sus scrofa, Phocoena sinus, and Lagenorhynchus albirostris have 1 disordered region within 35-residues Lfampin region not in 17-residues Lfampin Table 4. Although Equus asinus has 2 disordered regions but neither of them is within 17-residues Lfampin and both are found in 35-residues Lfampin region Table 4. Another unexpected data was those of Bos indicus, Bos grunniens, and Bos taurus, which have identical sequences for 17-residues Lfampin and 35-residues Lfampin region, and consequently the same number of disordered regions within 17-residues Lfampin and 35-residues Lfampin region as well as overall percent of disorder of 2 and 51.43%, respectively Table 4. The highest overall percent of disorder was for Capra hircus Lfampin (62.86%), whereas the lowest overall percent of disorder was for Homo sapiens (5.71%) Table 4.
The intrinsic disorder data of PONDR for camel Lfampin obtained in this study totally disagree with our previous report for camel Lf as well as Lfcin, which revealed that two long regions of camel lactoferrin including Lfcin sequence have greater disorder than the corresponding regions in both human and bovine proteins and peptides leading to superior antibacterial effects of camel protein and Lfcin as confirmed experimentally [3].

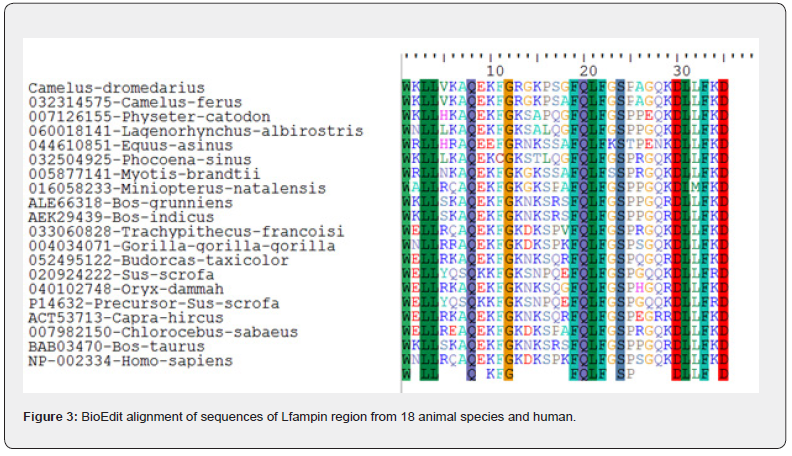
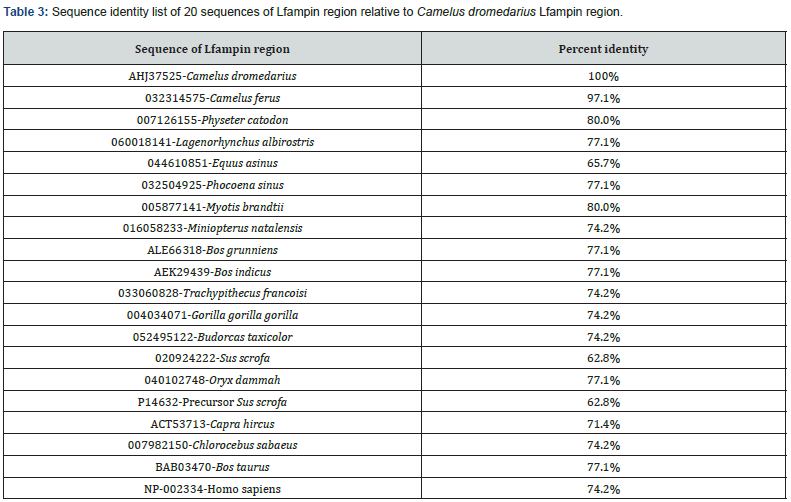
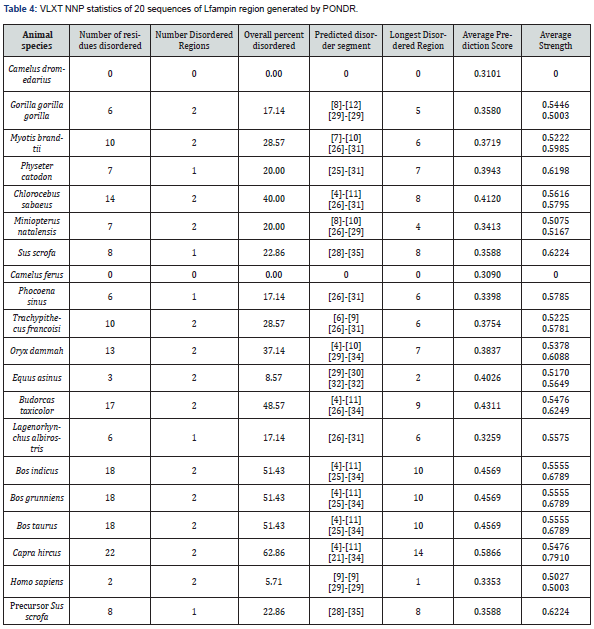
Figure 5 displayed results of prediction of disorder propensity and binary disorder in 20 sequences of Lfampin region from 18 animal species and human by flDPnn Server. These results of flDPnn disorder propensity vary significantly among analyzed sequences of Lfampin region. Distribution of intrinsic disorder in bioactive proteins/peptides as well as disorder propensity can identify their source such as animal milk source in this case. could identify animal meat source depending on the intrinsic disorder distribution peculiarities in mitochondrial cytochrome b amino acid sequences. They showed that discrimination between meat of avian and animal species could be predicted from the proportions of Ser-Pro-Ala, and Leu-Ile in mitochondrial cytochrome b amino acid sequences [33].
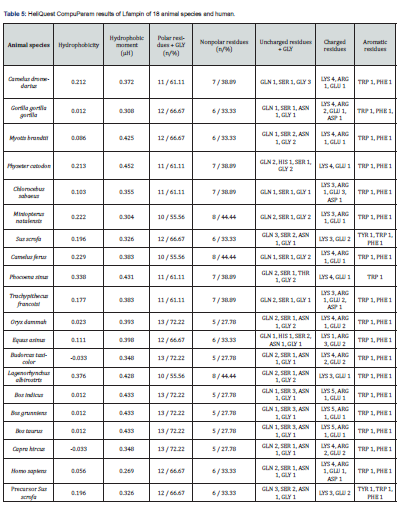
Enhancing the net positive charge of antimicrobial peptides such as Lfampin, i.e., improving cationicity of the peptide will significantly enhance its antibacterial effects or its antimicrobial effects in general. Additionally, predicting net positive charge of peptide can anticipate its antimicrobial effect and its potential application in feed or food, and as a peptide drug. HeliQuest CompuParam results of Lfampin of 18 animal species and human in Figure 6 and Table 5 demonastrated that Chlorocebus sabaeus Lfampin has zero net positive charge, while Bos indicus, Bos grunniens, and Bos taurus peptides have the highest net charge (=5). These net charge results of Bos taurus Lfampin in this work disagree with corresponding results of Bos taurus Lfcin we previously reported [34]. In our previous study, camel Lfcin has superior net charge over Bos taurus Lfcin and displayed greater antibacterial effects as confirmed experimentally [34].
Furthermore, Sus scrofa and Trachypithecus francoisi Lfampin have a net positive charge of only 1 Figure 6. Human Lfampin has a lower net positive charge (=3) relative to that of Bos taurus (=5), a result that agrees with that reported who increased the net positive charge of human Lfampin to enhance its Candidacidal and antibacterial effects [10].
Conclusion
Analysis of amino acid sequences, hydrophobicity, and charge characteristics of primary structure, number of disordered regions, overall percent of disorder, and disorder propensity of Lfampin from 18 different animal species and human recommends Bos grunniens (domestic yak Lfampin), Bos indicus (Zebu cattle Lfampin), and Bos taurus (cattle Lfampin) as candidates for developing therapeutic peptides either alone or linked to Lfcin in a chimeric peptide. These data should be experimentally validated. The absence of intrinsic disorder in camel Lfampin can be applied in identification of milk of this animal. Capra hircus (Goat Lfampin) with a net positive charge of 4 and 62.86% overall percent of disorder needs further investigation as a novel therapeutic peptide.
Conflict of Interest
None
References
- Van der Kraan MIA, Groenink J, Nazmi K, Veerman ECI, Bolscher JGM, et al. (2004) Lactoferrampin: a novel antimicrobial peptide in the N1-domain of bovine lactoferrin. Peptides 25(2): 177-183.
- Caccavo D, Pellegrino NM, Altamura M, Rigon A, Amati L, et al. (2005) Antimicrobial and immunoregulatory functions of lactoferrin and its potential therapeutic application. J Endotoxin Res 8(6): 403-417.
- Redwan EM, El Baky NA, Al Hejin AM, Baeshen MN, Almehdar HA, et al. (2016) Significant antibacterial activity and synergistic effects of camel lactoferrin with antibiotics against methicillin-resistant Staphylococcus aureus (MRSA). Research in Microbiology 167(6): 480-491.
- Almehdar HA, Abd ElBaky N, Mattar EH, Albiheyri R, Bamagoos A, et al. (2023) Exploring the mechanisms by which camel lactoferrin can kill Salmonella enterica serovar typhimurium and Shigella sonnei. PeerJ 11: e14809.
- Bruni N, Capucchio MT, Biasibetti E, Pessione E, Cirrincione S, et al. (2016) Antimicrobial activity of lactoferrin-related peptides and applications in human and veterinary medicine. Molecules 21(6): 752.
- Bellamy W, Takase M, Yamauchi K, Wakabayashi H, Kawase K, et al. (1992) Identification of the bactericidal domain of lactoferrin. Biochim. Biophys Acta 1121(1-2): 130-136.
- Gifford JL, Hunter HN, Vogel HJ (2005) Lactoferricin: a lactoferrin-derived peptide with antimicrobial, antiviral, antitumor and immunological properties. Cell Mol Life Sci 62(22): 2588-2598.
- Vogel HJ, Schibli DJ, Jing WG, Lohmeier Vogel EM, Epand RF, et al. (2002) Towards a structure–function analysis of bovine lactoferricin and related tryptophan- and arginine-containing peptides. Biochem Cell Biol 80(1): 49-63.
- Bradshaw JP (2003) Cationic antimicrobial peptides: issues for potential clinical use. Bio Drugs 17(4): 233-240.
- Haney EF, Nazmi K, Lau F, Bolscher JGM, Vogel HJ (2009) Novel lactoferrampin antimicrobial peptides derived from human lactoferrin. Biochimie 91(1): 141-154.
- Tang Z, Yin Y, Zhang Y, Huang R, Sun Z, et al. (2009) Effects of dietary supplementation with an expressed fusion peptide bovine lactoferricin–lactoferrampin on performance, immune function and intestinal mucosal morphology in piglets weaned at age 21 d. British Journal of Nutrition 101: 998–1005.
- Xie W, Song L, Wang X, Xu Y, Liu Z, et al. (2021) A bovine lactoferricin-lactoferrampin-encoding Lactobacillus reuteri CO21 regulates the intestinal mucosal immunity and enhances the protection of piglets against enterotoxigenic Escherichia coli K88 challenge. Gut Microbes 13(1): 1956281.
- Sarhadi H, Jahandar MH, Tanhaeian A (2020) Evaluation of antibacterial properties of chimeric bovine lactoferrin peptide for inhibition of food and plant pathogens. Int J Infect 7(2): e104594.
- Tanhaeian A, Mirzaii M, Pirkhezranian Z, Sekhavati MH (2020) Generation of an engineered food-grade Lactococcus lactis strain for production of an antimicrobial peptide: in vitro and in silico evaluation. BMC Biotechnology 20(1): 19.
- Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ (2000) Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform 11: 161-171.
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life J Mol Biol 337: 635-645.
- Shimizu K, Muraoka Y, Hirose S, Tomii K, Noguchi T (2007) Predicting mostly disordered proteins by using structure-unknown protein data. BMC Bioinformatics 8: 78.
- Garner E, Cannon P, Romero P, Obradovic Z, Dunker AK, et al. (1998) Predicting disordered regions from amino acid sequence: Common themes despite differing structural characterization. Genome Inform Ser Workshop Genome Inform 9 :201-213.
- Yang ZR, Thomson R, McNeil P, Esnouf RM (2005) RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 21(16): 3369-3376.
- Linding R, Russell RB, Neduva V, Gibson TJ (2003) GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res 31: 3701-3708.
- Li X, Romero P, Rani M, Dunker AK, Obradovic Z (1999) Predicting protein disorder for N-, C-, and internal regions. Genome Inform Ser Workshop Genome Inform 10: 30-40.
- Jones DT, Ward JJ (2003) Prediction of disordered regions in proteins from position specific score matrices. Proteins 53(Suppl 6):573-578.
- Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT (2004) The DISOPRED server for the prediction of protein disorder. Bioinformatics 20(13): 2138-2139.
- Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, et al. (2003) Protein disorder prediction: implications for structural proteomics. Structure 11: 1453-1459.
- Obradovic Z, Peng K, Vucet S (2003) Predicting intrinsic disorder from amino acid sequence. Proteins 53(Suppl 6): 566-572.
- Dosztanyi Z, Csizmok V, Tompa P, Simon I (2005) The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol 347(4): 827-839.
- Uversky VN, Gillespie JR, Fink AL (2000) Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins 41(3): 415-427.
- Garbuzynskiy SO, Lobanov MY, Galzitskaya OV (2004) To be folded or to be unfolded? Protein Sci 13: 2871-2877.
- Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, et al. (2005) Comparing and combining predictors of mostly disordered proteins. Biochemistry 44(6): 1989-2000.
- Tamura K, Stecher G, Kumar S (2021) MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 38: 3022-3027.
- Hall TA (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp Ser 41: 95-98.
- Théolier J, Fliss I, Jean J, Hammami R (2014) MilkAMP: A comprehensive database of antimicrobial peptides of dairy origin. Dairy Sci. Technol 94: 181-193.
- Yacouba HA, Sadeka MA, Uversky VN (2017) On the potential of using peculiarities of the protein intrinsic disorder distribution in mitochondrial cytochrome b to identify the source of animal meats. Intrinsically Disordered Proteins 5(1): e1264350.
- El Baky NA, Elkhawaga MA, Abdelkhalek ES, Sharaf MM, Redwan EM, et al. (2021) De novo expression and antibacterial potential of four lactoferricin peptides in cell-free protein synthesis system. Biotechnology Reports 29: e00583.






























