SDS-PAGE Electrophoresis of Ocypode rotundata Miers, 1882 and O. ceratophthalma Pallas, 1772 using Muscle Tissue along the Coast of Pakistan
Sahir Odhano1*, Noor US Saher2, Michael S Rosenberg3 and Mustafa Kamal4
1Department of Fisheries and Aquaculture, Shaheed Benazir Bhutto University of Veterinary and Animal Sciences, Pakistan
2Centre of Excellence in Marine Biology, University of Karachi, Pakistan
3Center for Biological Data Science, Virginia Commonwealth University, Richmond, Virginia, USA
4Department of Biotechnology, University of Karachi, Karachi, Pakistan
Submission:September 15, 2022; Published:September 29, 2022
*Correspondence author: Sahir Odhano, Department of Fisheries and Aquaculture, Shaheed Benazir Bhutto University of Veterinary and Animal Sciences, Pakistan Oceanogr
How to cite this article: Sahir O, Noor US S, Michael S R, Mustafa K. SDS-PAGE Electrophoresis of Ocypode rotundata Miers, 1882 and O. Ceratophthalma Pallas, 1772 using Muscle Tissue along the Coast of Pakistan. Oceanogr Fish Open Access J. 2022; 15(3): 555911. DOI: 10.19080/OFOAJ.2022.15.555911
Abstract
Electrophoretic studies for the identification of ghost crabs (Ocypode rotundata and Ocypode ceratophthalma) using the SDS-PAGE were performed on protein patterns along the Sandspit coastal areas. The muscle tissues were used to estimate the molecular mass of proteins through SDS-PAGE electrophoresis. Electrophoretic settings were standardized to use 10% acrylamide resolving gel and 5% acrylamide stacking gel. A discontinuous buffer system was used following the protocols of Laemmli (1970) to observe the relative mobility and molecular weight of proteins. In O. rotundata total of 6 protein bands were resolved among them band-5 was found similar in size to the marker used (BSA = 66 kDa MW). Whereas in O. ceratophthalma total of 8 protein bands were resolved among them three protein bands (band-6, band-7, and band-8) were found to be higher in size than the molecular marker. Both species revealed four protein bands smaller than BSA ranging from 20 kDa to 40 kDa MW. The smaller-sized protein bands were suspected to be myosin light chain protein bands, sarcoplasmic calcium-binding proteins (SCBP), tropomyosin, and arginine kinase. Results showed that both species of ghost crabs are distinct from each other through their banding pattern and their relative mobility. The current study revealed the possible efficacy of SDS-PAGE for the identification of ghost crab species by using the standard molecular marker. However, few molecular proteins may also be used as markers for species identification. Therefore, SDS-PAGE may also be used for species-specific protein identification and linked to immune-analytic techniques.
Keywords: Muscle Tissue, Ocypode; O. rotundata; O. ceratophthalma; Ghost crabs; SDS-PAGE; BSA; Sandspit, Karachi
Introduction
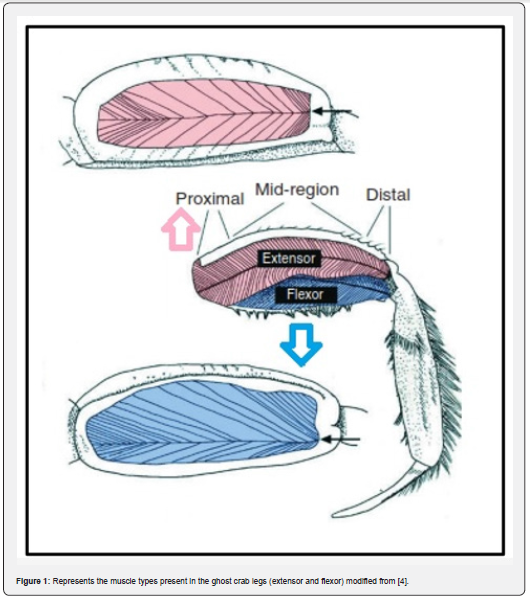
Ghost crabs are the most active, swift running, and aerobically fit crustacean species. These crabs are semi-terrestrial species and known as the fastest running invertebrates. They belong to the genus Ocypode Weber, 1795 (‘swift-footed’) family Ocypodidae. Their body muscles allow them to run swiftly at the maximum speed of 3-4 m/s [1-3] and can walk slowly and continuously for up to an hour [4]. There are two types of skeletal muscles involved in powering the crabs for fast running the extensors and flexors [4] (Figure 1). According to Priya et al. [3] these muscles receive enough supply of O2 through hemolymph for their running capabilities. The fiddler crabs also belong to the same family, but they are not capable of running too fast as compared to the ghost crabs.
Despite obvious differences between ghost crabs and other running vertebrates, the biochemical study revealed that the ghost crabs interestingly share many features of locomotion with other running vertebrates such as similar stride frequencies and trot to gallop transition [4,5]. The O2 supply to skeletal muscles occurs through the cardiovascular system to generate more energy for swift running. The physiological adaption for such a mechanism to support sustainable muscle power is partially known. Weibel & Hoppeler [6] studied vertebrate skeletal muscles and described the strong correlation between the aerobic capacity of whole animals and the volume of mitochondria within the skeletal muscles. There are many crustaceans who are active and highly alert due to the efficient supply of O2 to their skeletal muscles. But very little is known about skeletal muscle mechanism in invertebrates.
Electrophoretic studies are a widely used technique in tissues for protein analysis. Muscle tissues are the most suitable part for such a study to separate various peptide bands through their weight. Sodium Dodecyl Sulphate (SDS) is usually used in polyacrylamide gel electrophoresis (PAGE) to separate the protein according to their size and weight [7,8]. Therefore, SDS-PAGE is a technique used to isolate each peptide band, when current is applied and each band is observed in the gel [3,8-11].
Davis (1964) reported the method for separating serum proteins using polyacrylamide gel electrophoresis, and a significant advancement is observed in protein electrophoresis subsequently. Myofibril proteins are insoluble in PAGE electrophoresis and do not produce any results, therefore SDS was introduced in tissue sample and gel solution. This new approach helped the investigators resolve the issue and gave effective results [12,13]. Therefore, nowadays, SDS-PAGE is a simple technique to easily separate proteins through their molecular weight. SDS-PAGE is the most efficient, cost-effective technique, typically used to identify the molecular weight of proteins. SDS neutralizes the protein segment and denature it to estimate the protein weight easily
Materials and Methods
Study area
The specimens were collected randomly from the Sandspit (24.84, 66.91) sandy coastal area from 2014-2015 at the low tide. All the specimens were placed in tagged polyethene bags and brought to laboratory for electrophoretic study. Each specimen was sorted out through taxonomic identification keys [14,15]. O. rotundata specimen and O. ceratophthalma specimen were placed separately for muscle tissue extraction.
Tissue extraction
The muscle tissue sample was extracted (approx. 1g) from the enlarged cheliped of each individual species. The tissue sample was ground in a hand-held homogenizer in extraction buffer (Tris–citrate, 0.687M Tris and 0.157M Citrate pH 8.0) to homogenize the tissue prior to electrophoresis. The homogenized sample was centrifuged at 14500 rpm and the supernatant was poured into a new well-labelled Eppendorf tube for gel electrophoresis.
Solution preparation
The proteins present in tissue sample (Supernatant) were separated by SDS-PAGE vertical slab gel following the Laemmle (1970) protocols. A 10% acrylamide running gel (resolving) was prepared by using Tris-HCL buffer (pH 8.8), whereas a 5% spacer gel (stacking gel) was prepared by using Tris-HCL buffer (pH 6.8). Tris-glycine buffer (pH 8.5) was prepared as Tank buffer. The muscle tissue and A mixture of glycerol, tris-HCl buffer (pH 6.8), 10% SDS, and mercaptoethanol were homogenized through a centrifuge machine at 14500 rpm for the protein extraction.
Gel assembly casting
The glass plates of the electrophoresis apparatus were washed and carefully cleaned with ethanol and were fixed on the assembly using the spacers and clips. A resolving gel was prepared in a 50ml beaker ((3.2ml distilled water, 2.64ml 30% acrylamide, 2ml 1.5 M Tris, 0.5ml 10% SDS, 1ml 10% Ammonium persulfate (APS), 0.5μlml N, N, N, N N”-Tri methylamine triamine (TEMED) and the solution was poured into gel plate and waited for the gel to get polymerized (maximum 30 minutes). The resolving gel was poured about 2/3rd area of the gel plate and the rest of the area was filled with distilled water to level the resolving gel. When the resolving gel got polymerized and then distilled water was taken off and the stacking gel was prepared. The stacking gel was formed in a beaker (2.1ml distilled water, 0.5ml 30% acrylamide, 0.38ml of 1.0M Tris, 0.03ml 10% SDS, 0.03ml 10% APS, 0.003ml TEMED) the APS and TEMED were added just before pouring the upper cast of gel plates. Immediately the gel comb was inserted before the gel polymerized. After polymerization, the comb was removed, and wells were washed with distilled water.
Sample loading, staining and de-staining
A 20-μl of protein extract and 20-μl of sample buffer were mixed in well-labelled Eppendorf tubes and a 40-μl sample solution was formed. The 40-μl sample containing Eppendorf tubes was placed in hot boiling water for 2-3 minutes and then allowed to cool. Now, a 40-μl sample solution was loaded into the wells present in the staking gel. The reference standard Bovine Serum Albumin (BSA) was also loaded with each sample and assembly was allowed to run at 130 Volts (35 mA) for 3-5 hours or until the sample reached at the bottom of the gel. The gel was removed form glass plates carefully and stained with Coomassie brilliant blue R-250 for 24 hours. Then gel was replaced in de-staining solution for 30 minutes to observe the bands visibility. Process for replacing the de-staining solution was repeated if bands were not clearly visible. The de-staining solution was the mixture of methanol, acetic acid, and water with a ratio of 1:1:7 v/v.
When the gel was de-stained, and bands were observed on the gel then photographs were taken. The bands were observed manually, and their migration rate was recorded in a notebook for relative mobility analysis. The relative mobility (Rf) can be calculated by using the following formula:
Rf = distance migrated/dye front, length in gel
Results and Discussion
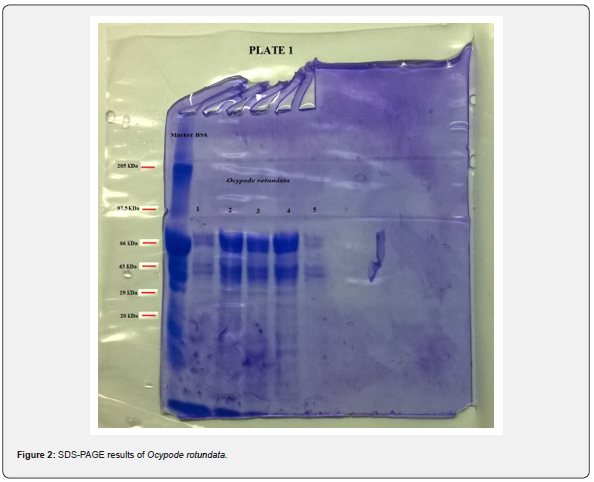
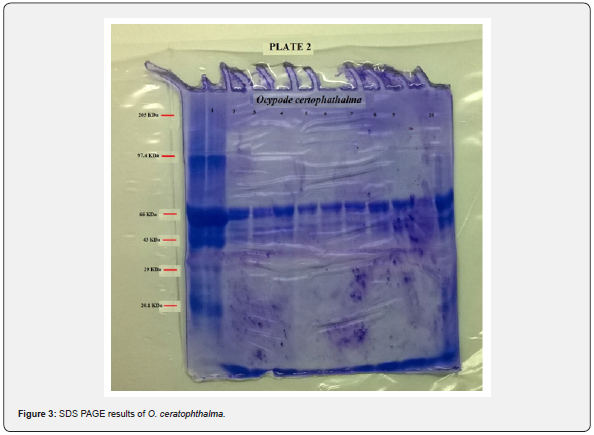
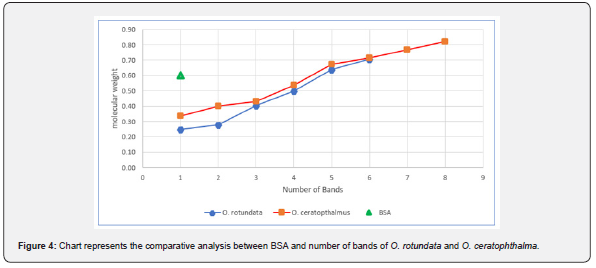
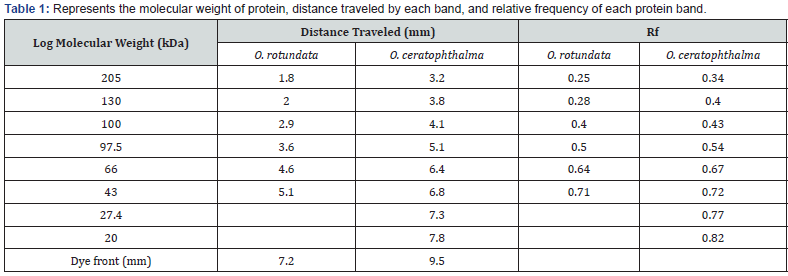
Electrophoretic analysis using SDS-PAGE of the muscle tissue was done. The present study revealed a total of 6 bands (peptides) resolved in the muscle tissue of O. rotundata (Figure 2), whereas O. ceratophthalma revealed a total of 8 bands (Figure 3). The four homologous proteins from band-3 to band-6 in both species were observed in the present investigation with a small fractional difference. The band-5 in both species found a similar size of BSA 66 kDa MW (Table 1). In the present study, several thick protein bands with strong intensity can be clearly seen in O. rotundata and O. ceratophthalma based on quantitative analysis with bands revealed 4 protein bands (band-1 to band-4) smaller than 66 kDa MW, whereas only one protein band (band-6) in O. rotundata and 03 peptide bands (band-6 to band-8) in O. ceratophthalma found larger in size than BSA (>66 kDa MW). The results of quantitative analysis of banding patterns using the SDS-PAGE can be seen in figure 4. The smaller-sized protein bands were suspected to be myosin light chain protein bands, sarcoplasmic calcium-binding proteins (SCBP), tropomyosin, and arginine kinase ranging between 20 kDa to 40 kDa MW comparing the present data with Arwani et al. [16]. Such results may also be compared with allergen proteins in white shrimp as categorized by the IUIS-International Union of Immunological Societies and WHO-World Health Organisation, 2021. Other brachyuran crabs such as Scylla serrata revealed thick with strong intensity bands ranging from 25 kDa to 65 kDa MW [17]. Arwani et al., [16] stated boiling protein extract can greatly reduce the intensity and number of proteins up to -36% in arginine kinase, -18% in myosin light chain, and increase the tropomyosin protein intensity up to +528% in mud crab [18]. Kim et al. [19] described that the reduced number of protein bands were likely to be degraded into smaller molecular weight protein bands. It is also possible that proteins that have a smaller resolution limit (less than 10% acrylamide) may travel faster and drop down into the buffer [16,20].
The muscle tissues collected from ghost crabs provided the best results with clear resolution of protein bands using SDSPAGE. The proteins present in muscle tissues can be used as marker proteins to help in the various shellfish species identification through the specific/unique protein bands present in each species’ muscle profile. The whole-body homogenates are less successful and produce distinct proteins bands as compared to the muscle tissues. Besides this, large number proteins may dilute the essential proteins and prevent a precise quantitative analysis of individual protein bands [21]. Dubey & Flynn [22] suggested that protein resolution is controlled by percentage of acrylamide as used in any study. Kitts et al. [21] recommended 12% acrylamide gels for greater resolution of bands for muscle tissues.
The SDS-PAGE is the tool to separate protein extract into specific protein bands. These bands provide sources of identifying differences between shellfish species [21]. Electrophoresis is a simple technique for species identification, and it requires less cost as compared to other species identification methods [23]. However, Scobbie and Maackie [24] argued that proteins get de-natured due to boiling therefore, this method is not suitable for species identification. Furthermore, due to the increasing demand for shellfish species in the food market, it is necessary to use reliable, sensitive methods to identify species to follow the food and labeling regulations [21]. Therefore, An et al., [25] and Taylor and Jones [26,27] suggested immunoassay methods such as Enzyme-linked immunosorbent assays (ELISA) are useful for shellfish species identification.
Conclusion
The use of SDS-PAGE in the current study is an effective method for identifying species differences in ghost crabs. The SDS-PAGE is also recommended for relative concentration, and molecular mass estimation of proteins since 1970, as it provides a higher resolution of protein bands [10,28-30]. A few soluble proteins may also be used as markers for species identification. This methodology provides necessary basic knowledge related to species-specific protein bands that focus on the opportunities for biotechnology as it is linked to immune-analytic techniques [31-39].
Acknowledgment
This research was partially supported by Pakistan Science Foundation, Project No. Bio-456. We are thankful to anonymous reviewers for their timely review of this article.
References
- Hafemann DR, Hubbard JI (1969) On the rapid running of ghost crabs (Ocypode ceratophthalma). Journal of Experimental Zoology 170(1): 25-31.
- Herreid CF, Full RJ (1988) Energetics and locomotion. In: Burggren WW, McMahon BR, (edn.), Biology of the land crabs, Cambridge: Cambridge University Press, UK, pp. 333-337.
- Priya MS, Kumarasamy P, Pugazhendi A, Ganapiriya V, Muthukumaravel K (2019) Electrophoretic Analysis of Reproductive Tissues and Haemolymph of the Marine Pebble Crab Leucosia Anatum (Herbst, 1783). International Journal of Pharmacy and Biological Sciences 9(1): 89-93.
- Perry MJ, Tait J, Hu J, White SC, Medler S (2009) Skeletal muscle fiber types in the ghost crab, Ocypode quadrata: implications for running performance. Journal of Experimental Biology 212(5): 673-683.
- Blickhan R, Full RJ (1987) Locomotion energetics of the ghost crab: II. Mechanics of the centre of mass during walking and running. Journal of experimental biology 130(1): 155-174.
- Weibel ER, Hoppeler H (2005) Exercise-induced maximal metabolic rate scales with muscle aerobic capacity. Journal of Experimental Biology 208(9): 1635-1644.
- Roy S, Kumar V (2014) A practical approach on SDS PAGE for separation of protein. International Journal of Science and Research 3(8): 955-960.
- Saher NU, Odhano S, Mustafa K (2017) Electrophoretic analysis for the separation of muscle protein of fiddler crabs of Pakistan. International Journal of Biology Research 5(2): 77-81.
- Khan JM, Qadeer A, Chaturvedi SK, Ahmad E, Abdul RSA, et al. (2012) SDS can be utilized as an amyloid inducer: a case study on diverse proteins. PloS one 7(1): e29694.
- Nowakowski AB, Wobig WJ, Petering DH (2014) Native SDS-PAGE: high resolution electrophoretic separation of proteins with retention of native properties including bound metal ions. Metallomics 6(5): 1068-1078.
- Yang S, Douglas TD, Ruia R, Medler S (2021) Hemolymph supply to locomotor muscles of the ghost crab Ocypode quadrata. Journal of Experimental Biology 224(13).
- Greaser ML, Yates LD, Krzywicki K, Roelke DL (1983) Electrophoretic methods for the separation and identification of muscle proteins [Meat]. In Proceedings Annual Reciprocal Meat Conference.
- Kresge N, Simoni RD, Hill RL (2006) SDS-PAGE to determine the molecular weight of proteins: the work of klaus weber and Mary Osborn. Journal of Biological Chemistry 281(24): 19-21.
- Sakai K, Türkay M (2013) Revision of the genus Ocypode with the description of a new genus, Hoplocypode (Crustacea: Decapoda: Brachyura). Memoirs of the Queensland Museum 56(2): 665-793.
- Shih HT, Ng PK, Davie PJ, Schubart CD, Türkay M, et al. (2016) Systematics of the family Ocypodidae Rafinesque, 1815 (Crustacea: Brachyura), based on phylogenetic relationships, with a reorganization of subfamily rankings and a review of the taxonomic status of Uca Leach, 1814, sensu lato and its subgenera. Raffles Bulletin of Zoology, p. 64.
- Arwani A, Palupi NS, Giriwono PE (2022) Effects of Different Heat Processing on Molecular Weight and Allergenicity Profile of White Shrimp (Litopenaeus vannamei) and Mud Crab (Scylla serrata) from Indonesian Waters. Squalen Bulletin of Marine and Fisheries Postharvest and Biotechnology 17(1): 13-22.
- Ar NI, My ZH, Rosmilah M, Noormalin A, Faizal B, et al. (2015) Identification of major and minor allergens of mud crab (Scylla Serrata). Medicine and Health 10(2): 90-97.
- Lasekan AO, Nayak B (2016) Effects of buffer additives and thermal processing methods on the solubility of shrimp (Penaeus monodon) proteins and the immunoreactivity of its major allergen. Food Chemistry 200: 146-153.
- Kim KBWR, Lee SY, Song EJ, Kim KE, Ahn DH (2011) Effect of heat and autoclave on allergenicity of porcine serum albumin. Food Science and Biotechnology 20(2): 455-459.
- Yadzir ZHM, Misnan R, Bakhtiar F, Abdullah N, Murad S (2015) Tropomyosin, the major tropical oyster Crassostrea belcheri allergen and effect of cooking on its allergenicity. Allergy, Asthma & Clinical Immunology 11(1): 1-6.
- Kitts DD, Shum MLP, Smith DS (2020) Species identification of shellfish using SDS-Page electrophoresis. In: Seafood safety, processing, and biotechnology. CRC Press, pp. 235-241.
- Dubey S, Flynn E (1998) Estimating protein molecular weights using SDS-PAGE. FOCUS Invitrogen 20: 24-25.
- Sotelo CG, Piñeiro C, Gallardo JM, Pérez MRI (1993) Fish species identification in seafood products. Trends in Food Science & Technology 4(12): 395-401.
- Scobbie AE, Mackie IM (1988) The use of sodium dodecyl sulphate polyacrylamide gel electrophoresis in fish species identification–a procedure suitable for cooked and raw fish. Journal of the Science of Food and Agriculture 44(4): 343-351.
- An H, Klein PA, Kao KJ, Marshall MR, Otwell WS, et al. (1990) Development of monoclonal antibody for rock shrimp identification using enzyme-linked immunosorbent assay. Journal of agricultural and food chemistry 38(11): 2094-2100.
- Taylor WJ, Jones JL (1992a) An immunoassay for verifying the identity of canned sardines. Food and Agricultural Immunology 4(3): 169-175.
- Taylor WJ, Jones JL (1992b) An immunoassay for distinguishing between crustacean tailmeat and white fish. Food and Agricultural Immunology 4(3): 177-180.
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259): 680-685.
- Walker JM (2002) SDS polyacrylamide gel electrophoresis of proteins. In: The protein protocols handbook. Humana press pp. 61-67.
- Gallagher SR, Sasse J (2008) Staining proteins in gels. Current Protocols Essential Laboratory Techniques 1: 7-4.
- Bertolini MJ, Tankersley DL, Schroeder DD (1976) Staining and destaining polyacrylamide gels: a comparison of coomassie blue and fast green protein dyes. Analytical Biochemistry 71(1): 6-13.
- Chrambach A, Reisfeld RA, Wyckoff M, Zaccari J (1967) A procedure for rapid and sensitive staining of protein fractionated by polyacrylamide gel electrophoresis. Analytical biochemistry 20(1): 150-154.
- Sasse J, Gallagher SR (2008) Detection of proteins on blot transfer membranes. Current protocols in immunology 83(1): 8-10.
- Gallagher SR (2008) SDS‐polyacrylamide gel electrophoresis (SDS‐PAGE). Current protocols essential laboratory techniques 1: 7-3.
- Giulian GG, Moss RL, Greaser M (1983) Improved methodology for analysis and quantitation of proteins on one-dimensional silver-stained slab gels. Analytical biochemistry 129(2): 277-287.
- Gorovsky MA, Carlson K, Rosenbaum JL (1970) Simple method for quantitive densitometry of polyacrylamide gels using fast green. Analytical biochemistry 35(2): 359-370.
- Weber K, Osborn M (2006) SDS-PAGE to determine the molecular weight of proteins: The work of Klaus Weber and Mary Osborn-The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis (reprinted from J. Biol Chem (1969) 244: 4406-4412). Journal of Biological Chemistry 281(24): 19-21.
- Laemmli UK, Quittner SF (1974) Maturation of the head of bacteriophage T4: IV. The proteins of the core of the tubular polyheads and in vitro cleavage of the head proteins. Virology 62(2): 483-499.
- Saher NU (2008) Population dynamics and biology of fiddler crab in the mangrove area of Karachi coast (Doctoral dissertation, University of Karachi, Karachi).






























