The Genetic Research Methods and Its Role in Aquaculture On Indonesia
Bruri Melky Laimeheriwa1,2*
1Department of Aquatic Resources Management, Faculty of Fisheries and Marine Science, Maritime and Marine Science Center for Excellence of Pattimura University Ambon, Indonesia
Submission: January 09, 2018; Published: September 14, 2018
*Correspondence author: Department of Aquatic Resources Management, Faculty of Fisheries and Marine Science, Maritime and Marine Science Center for Excellence of Pattimura University Ambon, Indonesia.
How to cite this article: Bruri Melky L. The Genetic Research Methods and Its Role in Aquaculture On Indonesia. Oceanogr Fish Open Access J. 2018; 8(3): 555738. DOI:10.19080/OFOAJ.2018.08.555738
Abstract
Successful development of aquaculture is largely determined by the seed supply of both the quality and quantity of the seed. Seed quality is influenced by the quality of the mother (genetic factors) and environmental factors (water quality, food, and disease). Improved parent quality through the improvement of both qualitative and quantitative genetic traits that can be done through the selection of traits or characteristics of prospective mothers, as well as avoiding the occurrence of inbreeding that causes heritabilities decline. The purpose of writing to review the results of genetic studies that have been applied in the field of aquaculture in Indonesia. In this article, the study included the following: basic genetic concepts and theories, genetic research methods, and their roles and functions in the field of aquaculture.
Keywords: Research method; Aquaculture; Character; Gynogenesis; Androgenesis; Heritability; Breeding; Genetic
Introduction
Aquaculture is any form of activity to produce biota or aquatic organisms in a controlled environment and in nature in order to obtain production (output) and benefits (outcomes). Activities carried out in the sea, brackish waters, fresh water, including the general waters of the lake, reservoirs, and rivers. In principle, aquaculture activities are to control growth, death, and reproduction.
Observing the status and potential of our nation’s aquaculture, although overall national fishery production is still dominated by capture fisheries, the contribution of aquaculture has a much higher production growth compared to capture fisheries. Data from the Ministry of Marine Affairs and Fisheries 2014 shows that the contribution of aquaculture to national production increased from 18.05 percent in 2009 to 20.56 percent in 2013. In contrast, the contribution of capture fisheries decreased from 81.95 percent in 2009 to about 79.44 percent in 2013 (CTF Statistics, 2014). This gives the impression that aquaculture in the future will play an increasingly important role, but it certainly must be accompanied by some notes of improvement of the weakness that has been found.
Domestic and world demand for fishery products continues to increase as population increases and human awareness of the benefits of healthful and educated fish increases. The capability of producing fishery products from capture fisheries activities at the global level is maximum of 100 million tons per year (FAO, 2014) and national 16.4 million tons per year. Now its catch quantity tends to decrease.
The potential of aquaculture production owned by Indonesia is about 57.7 million tons per year (the largest in the world). While in 2013 the total production of national aquaculture is only 1.5 million tons (2.6 percent). Thus, if aquaculture is more excavated and empowered through change and improvement, it is not impossible that the future will become a mainstay of the economy and increase the effect of a very large domino again. In the sense of providing opportunities for economic growth, job creation, and reducing poverty.
More than 90 percent of the world’s aquaculture production comes from Asia, so it is only natural that Asian countries are the most likely to attract foreign exchange from aquaculture business. Indonesia has the opportunity to become a world leader in aquaculture as well as a mainstay of the economy. Moreover, the government’s attention to this sector is increasingly evident with the big vision of making Indonesia sovereign in the maritime field by making Indonesia as the maritime axis of the world.
Successful development of aquaculture is highly determined, among others: by the supply of seeds that include the quality and quantity factors of seed. Seed quality is influenced by the quality of the mother (genetic factors) and environmental factors (water quality, food, and disease). The quality of the parent as a genetic quality factor has an influence on the quality of the seeds in terms of increasing the number of seeds, rapid growth, and resistance to environmental changes (disease and water quality). Improving the quality of the mother through the improvement of both qualitative and quantitative genetic traits that can be done through selection of traits or characters of prospective mothers. In addition, the management of the parent to avoid the occurrence of inbreeding that causes decreased heritability the parent.
This manuscript aimed to review the results of genetic studies that have been applied in the field of aquaculture. In this article, the study includes basic genetic concepts and theories, genetic research methods, and the role and function of genetic analysis in aquaculture.
The Basic Concepts and Theories of Genetics
Basic concept of heredity
Although people usually establish genetics beginning with the rediscovery of the manuscript of an article written by Gregor Mendel in 1900, genetics as “the science of inheritance” or heredity has been known since prehistoric times, such as the domestication and development of various breeds of cattle and cultivars. People have also known the effects of crossing and close marriage and made a number of procedures and regulations about it since before genetics stood as an independent science. Genealogy of the disease in the family, for example, has been reviewed before. However, this practical knowledge does not explain the cause of the symptoms.
The popular theory of inheritance adhered to in that period is the theory of inheritance: one inherits a flat mixture of attributes brought by the elderly, especially of the males for carrying sperm. Mendel’s results show that this theory is not applicable because the properties are carried in combinations carried by typical alleles, rather than mixed blends. Another related opinion is Lamarck’s theory: the traits that the elder inherited in his life be passed on to his son. This theory is also broken with Mendel’s explanation that the nature brought by the gene is not influenced by the experience of the individual who inherits that trait. Charles Darwin also gave an explanation of the pangenesis hypothesis and later modified by Francis Galton. In this view, the body’s cells produce particles called gemmules that will be collected in the reproductive organs before fertilization occurs. Thus, every cell in the body has a contribution to the properties that its offspring.
The basic concept of Gen
Genes (from the Dutch: genes) are the inheritance units of nature for living organisms. The physical form is a sequence of DNA encoding a protein, polypeptide, or an RNA that has a function for the organism that has it. Modern gene boundaries are a particular location in the genome that is related to inheritance properties and can be associated with functions as regulators (controllers), transcription goals, or other functional roles. The use of “genes” in everyday conversations (eg “smart genes” or “hair gene genes”) are often meant for alleles: the choice of variations available by a gene (Figure 1). Although the allele’s expression can be similar, people more often use the term allele for expression of genes that are phenotypically different. The genes are inherited from one individual to their offspring through a process of reproduction, together with the DNA that carries it. Thus, information that maintains the integrity of the form and function of an organism’s life can be maintained.
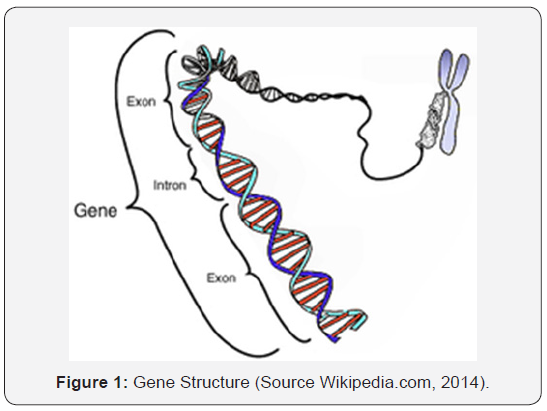
At that time DNA has been discovered and is known to reside only on chromosomes (1869), but people have not yet realized that DNA is linked to genes. Through Oswald Avery’s study of Pneumococcus bacteria (1943), and Alfred Hershey and Martha Chase (publication 1953) with T2 bacteriophage virus, did people know that DNA is a genetic material. In the 1940s, George Beadle and Edward Tatum experimented with Neurospora crassa. From these experiments, Beadle and Tatum can draw hypotheses that genes encode enzymes, and they conclude that one gene synthesizes one enzyme (one gene-one enzyme theory). A few decades later, it was discovered that genes encode proteins that not only function as enzymes and some proteins are composed of two or more polypeptides. With these findings, Beadle and Tatum’s opinion, one gene-one enzyme theory, has been modified into the theory of one gene-one polypeptide (one gene-one polypeptide theory).
In eukaryotic cells, the gene consists of the transcriptional initiation regulatory domain, comprising the following: GCCACACCC series, ATGCAAAT, GC boxes, CCAAT boxes and TATA boxes, introns, and exons that are encoded protein areas that can be transcribed overlapping or nonoverlapping. For example, in a code with three series of nucleotides (Tripcode codons) AUU GCU CAG, it can be read nonoverlapping as AUU GCU CAG or read overlapping as UGC UUG UGC GCU CUC CAG. Although in about 1961, it has been found that amino acids coded by codons by nonoverlapping have found different proteins transcribed with shifting overlapping codons, as well as the final regulatory domain of transcription.
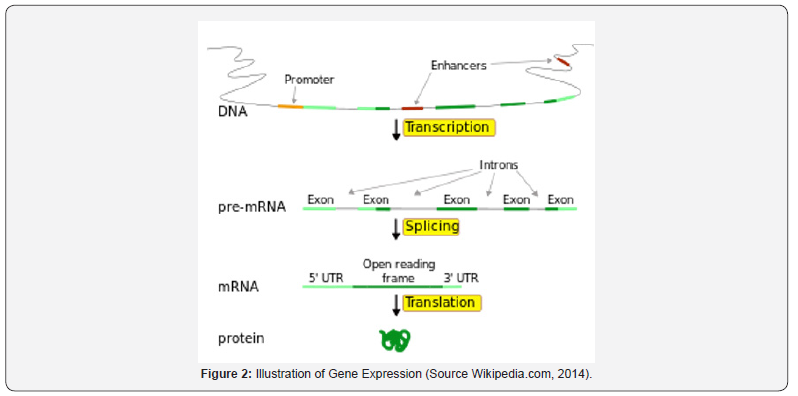
Gene expression is the process by which the information codes present in the gene are converted into proteins that operate only within the cell. Gene expression consists of two stages: the process of making RNA copies (transcription) and polypeptide synthesis processes specifically in the ribosome (translational). The process of transcribing DNA into mRNA and translating mRNA into a polypeptide is called the central dogma. The central dogma applies to prokaryotes and eukaryotes. However, in eukaryotes, there is an additional stage that occurs between transcription and translation called the pre-mRNA stage. The pre-mRNA stage is to select the mRNA to be sent out of the nucleus to translate in the ribosome. The exon is the mRNA to be sent out of the nucleus to be translated, while the introns are the mRNAs that will remain in the nucleus because it is likely that the mRNA will form a protein that is not functional if it is translated. Intron will then break down again to form a new mRNA chain. It is also known that some of the so-called mutations can occur in the process of expression of this gene (Figure 2).
The basic concept of genotype
Genotype (literally “gene type”) is the term used to express the genetic state of an individual or a group of individual populations. Genotypes may refer to the genetic state of a locus as well as the entire genetic material carried by the chromosome (genome). Genotypes can be homozygous or heterozygous. Once people can transfer gene, there is also the use of the term hemizygote.
In Mendel’s genetics (classical genetics), genotypes are often denoted by pairs of letters; such as AA, Aa, or B1B1. The same pair of pairs indicates that the individual symbolized is homozygous (AA and B1B1), while the different letter pairs represent the heterozygous individual. A pair of letters indicates that this symbolized individual is diploid (2n). As a consequence, the individual tetraploid (4n) homozygote is denoted by AAAA, for example.
The basic concept of phenotype
The phenotype is a characteristic (both structural, biochemical, physiological, and behavioral) that can be observed from an organism regulated by genotype and its environment and interaction. The phenotypic definition includes the various levels of the gene expression of an organism. At the organism level, a phenotype is something that can be seen/ observed/measured, something of nature or character. In this stage, phenotypes such as eye color, weight, or resistance to a particular disease. At the biochemical level, the phenotype may be the content of certain chemical substances in the body. For example, blood sugar or protein content in rice. At the molecular level, the phenotype may be the amount of RNA produced or the detection of DNA or RNA bands in electrophoresis.
Phenotypes are determined in part by individual genotypes, in part by the environment in which the individual lives, time, and, in some traits, the interaction between genotype and environment. Phenotypic observations can be simple (usually flower color) or very complicated to require special tools and methods. However, due to the genetic expression of a gradual genotype from the molecular level to the individual level, there is often a link between a number of phenotypes at different levels.
The basic concept of inheritance of qualitative and quantitative properties
So far the discussion of a phenotype is assumed to describe its genotype. Phenotypes of such traits are easy to distinguish, for example, the color of the bean husk is alternatively white or gray, the plant is high or low, so it is easy to distinguish. Such properties are known as discontinuous traits. In such traits, there is a simple relationship between genotype and phenotype. In many cases a genotype produces only one kind of phenotype, otherwise, a phenotype is the result of the activity of a genotype. Nevertheless, we have come to understand that the relationship between genotype and phenotype is influenced by phenomena, such as expressivity, penetration, and pleiotropy. In other words, a genotype can produce different phenotypes because the genotype interacts with its environment during the growth and development process. The properties such as fish weight, fish length and protein content show a wide range of phenotypes.
The properties that indicate such a range of phenotypic diversity are often called continuous traits. On discontinuous properties have a discrete distribution, whereas continuous properties have a continuous distribution, so it must be expressed quantitatively. Such properties are often called quantitative properties. Continuous properties have a continuous range of phenotypes as well. To examine the inheritance of the continuous/quantitative nature it is necessary to see why a trait has various phenotypes. Quantitative properties can occur in various ways. Generally, the phenotype range occurs because different genotypes exist in an individual group (population). This is common when a character is controlled by many loci. For example, if a locus with two alleles per locus controls a trait, then there are three possible genotypes namely AA, Aa, aa. For two loci with two alleles per locus, there will be 9 genotypes (32), eg AABB, AABb, Aabb, AaBB, AaBb, Aabb, aaBB, aaBb, and aabb. Similarly, the increase in the number of loci as much as n, then the genotype formed as much as 3n. if there are more than two alleles for each locus it will produce much more genotype.
Thus, the more the locus that controls a trait, the more likely genotypes may be formed. The properties controlled by many such loci are called polygenic or quantitative properties. polygenic or quantitative properties. The basic difference between qualitative and quantitative properties involves the number of genes that contribute to the phenotype variability and the degree to which the phenotype can be modified by environmental factors. Quantitative properties can be governed by many genes (perhaps 100 to 100 or more), each contributing to the phenotype so little that its individual influences cannot be detected by Mendel’s methods. Such genes are called polygenic.
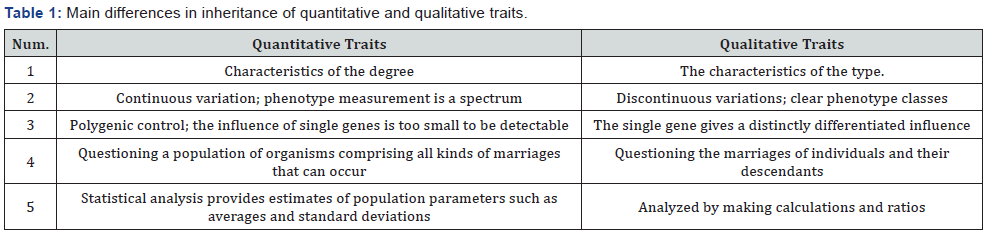
Below is presented a summary of some major differences between quantitative and qualitative genetics (Table 1). In the inheritance of quantitative properties, the concept of polygenic (polygenes, meaning “many genes”) is used to explain the formation of quantitative traits. Ronald Fisher can explain that the quantitative nature is formed of many genes with little influence, each of which segregates according to Mendel’s theory. Because of its small effect, the phenotype regulated by these genes can be affected by the environment. Nevertheless, Fisher’s explanation still places the “genes” that govern quantitative properties as abstract because they are only concepts. The proof of the existence of genes that regulate quantitative properties begins to open up after the availability of many genetic markers to enable people to create genetic maps that can reach most of the chromosomes. Genetic markers are used to indicate the allelic situation in a particular chromosome part. Allele variations on a marker become genotypes for chromosomes or clusters.
Basic concepts of inheritance or heritability
According to Nasir [1] heritability is the proportion of genetic variation to the total amount of genetic variation coupled with the variety of environment, in other words, heritability is the proportion of the genetic variation to the phenotype variety for a particular character. There are two heritability values known in plant breeding that are heritability in the broad sense and heritability in the narrow sense [2].
The value of heritability in the broad sense takes into account the total genetic variation in relation to phenotype diversity. In this case, the genotype is considered a unit in relation to the environment. While narrow heritability is the focus of attention, the diversity attributed to the role of additive genes is part of the total genetic diversity. Based on this explanation it can be understood that the value of heritability in the narrow sense will never be greater than the value of heritability in a broad sense for a particular character [2].
Heritability parameters involve all types of gene action and therefore form a broad heritability estimate. In the case of perfect dominance, if a gamete containing the dominant active allele A2 combines with a gamete containing a zero allele A1, the resulting phenotype may consist of two units. When two A2 gametes combine, the phenotype results will consist of two units. Conversely, if genes that have no dominance (additive genes) are involved, then the A2 gametes will add one unit to the phenotype of the resulting zygote, regardless of the contribution of the allele of the gamete to which it combines. Thus, only the genetic additive component of the variance has predictable quality, which is in the formulation of breeding roles. Heritability in this narrower sense is the ratio of genetic additive variance to the phenotype variance.
Methods of Genetic Research
Method analysis of phenotype performance
Phenotypic performance analysis includes morphological and morphometric data. Measured morphometric data include weight, length, width, height, shell thickness, area, circumference, etc. The morphometric ratio/index analysis is performed to obtain the coefficient of interpersonal performance within a population by calculating the distance coefficient (Eucleudian value), the coefficient of similarity and coefficient of variation. The coefficient values are mapped in the dendrogram to see whether there are similarities and differences between individuals in the ring snail population [3].
Biochemical analysis method
The measured biochemical performance was the analysis of enzyme (allozyme), protein, amino acids using starch gel electrophoresis. The analysis of the buffer migration rate and the staining procedure follow the Sugama method et. al. Fitness test for Hardy-Weinberg equilibrium and differences between populations using GENEPOP and GenStat software [4]. The interpretation of locus and allele identification follows the Allendorf & Utler method [5]. The allele’s naming is based on the relative distance of the migration of the allele to the most commonly encountered allele (mode) coded 100. The data is interpreted to obtain allelic frequency, the degree of polymorphism, heterozygosity and genetic distance.
Discriminant analysis was used to see the significant differences between groups determined by observed morphometric. The measured results are relativized using standard deviations to avoid the diversity of sizes and possibly different ages. Next will be grouped morphological characters that most characterize the difference. The influence of the interaction of fellow characters measured resulted in the depreciation of these characters. This component is also used to see the most powerful influence on the formation of a population. This analysis uses Minitab program version 17 and SPSS ver. 20 program based on Principal Component Analysis (PCA). PCA analysis aims to interpret most of the information contained in a data matrix into graphs and to obtain maps of population distribution and similarity values within and outside the group. The results obtained are shown in dendrogram form.
RNA and DNA ratio analysis methods
Total DNA and RNA extracts follow the standard method developed by Farajallah et al. [6]. Amplification and visualization of DNA and RNA using PCR and electrophoresis methods with primers tailored to the sample. DNA sequencing and RNA from PCR using Sequenzer ABI377A Applied Biosystem. The DNA sequence and RNA are edited manually using Genetyx software, then aligned using Clustal W software [7]. Characterization of DNA sequences and their analysis using the maximum parsimony (MP) and Neighbor Joint (NJ) methods performed using MEGA software [8]. To find out the concentration of DNA and RNA measured by using Gene quant program. Results of DNA and RNA genomic concentrations of each sample were further calculated for determination of RNA-DNA ratio.
DNA marker methods
Some DNA markers that can be used to analyze genetic variation, the evolution of a plant, the linking of specific genes to specific characters, parent search, analysis of quantitative character loci, and revision of plant classification are Restriction Fragment Length Polymorphism (RFLP), Random Amplified Polymorphic DNA (RAPD), Amplified Fragment Length Polymorphism (AFLP), and microsatellite or Simple Sequence Repeat (SSR).
Restriction fragment length polymorphism (RFLP)
Research to exploit DNA polymorphisms in plant genomes by utilizing the technology of molecular markers continues to increase. Restriction Fragment Length Polymorphism (RFLP) is a molecular technique based on polymorphism caused by the substitution of the base (nucleotide), insertion, dilation, or translocation that may occur in the past. This technique utilizes specific restriction sites in the genome of an organism. Restriction enzyme or restriction endonuclease is an enzyme that cuts DNA strands on a sugar-phosphate framework without damaging a base, in the sequence it recognizes. The recognition sequence is often called the introduction site is a DNA sequence that places the restriction enzyme and cuts on the sequence. These enzymes can be ordered through companies engaged in biotechnology and molecular biology. The result of genomic cutting using certain restriction enzymes will result in differences in the length of the DNA fragment, which shows the distance from the enzyme restriction sites in an organism’s genome. The fragmented genome can then be analyzed for the purpose of the study. Through further analysis can be known whether the target sequence has been changed due to the substitution of the base (nucleotide), insertion, deletion, or translocation.
Random amplified polymorphic DNA (RAPD)
RAPD is widely used to analyze the diversity of genetic traits in a variety of studies with the consideration that, among others, it does not require a background knowledge of the genome to be analyzed, primers used universally (can be used for both prokaryotes and eukaryotes), capable of producing relatively unlimited characters, - the materials used are relatively cheaper, the preparation is easier, and yields faster results than other molecular analyzes. The RAPD method is able to detect the nucleotide sequence by using only one primer. The primer will bind to the single strand of the one genome and to the DNA strand of its partner in the opposite direction. As long as the primary attachment site remains at an amplified range in general no more than 5000 base pairs (bp), it will obtain amplification DNA products. With the use of RAPD that is relatively easy, inexpensive, and analyzed in DNA level, early selection can be performed on the properties of resistance to biotic and abiotic stress, quantitative characteristics, and duplication of collection crops can be avoided.
Microsatellite (simple sequence repeat, SSR)
Repeated recurrent DNA that has the highest variation in the plant genome is a recurrent sequence with a simple or short repetitive fragment. This fragment is known by the name minisatellite and microsatellite. Minisatellite is a repetitive DNA usually between 10-60 base pairs, which at the beginning of its discovery was widely applied to the human genome. While microsatellite (SSR) has fewer recurring units ranging from 1 to 6 base pairs, contained in very large quantities and spread in the genome, and is widely used in plants. Variations of these fragments are usually the result of changes in the number of copies of the original loop and are often categorized as a variable number of tandem repeats (VNTR). Because very high levels of polymorphism can be detected with this fragment, VNTR is recognized as a potent tool for fingerprinting and cultivar identification. This fragment can also be used to study the diversity between and intra-population, ecological studies, calculating genetic distance and studying plant evolution. The short sequence of DNA microsatellites with the clamp DNA sequences is conserved, allowing the primary design to amplify specific sites using PCR. If these primers are used to amplify certain SSR loci, then each primer will produce polymorphism in the form of a difference in amplification length known as SSLP (Simple Sequence Length Polymorphism). Each length represents one allele of a locus. Long differences occur because of the difference in the number of repeating units at specific SSR loci. The diversity of the number of replicates in microsatellite can be detected by electrophoresing the amplified DNA products in the standard gel sequence, which can separate the fragments that distinguish each nucleotide. Microsatellite DNA is present in large quantities and spreads in the genome. The general form of repetition of DNA microsatellite (SSR) is a simple repetition of two bases. Microsatellite DNA (SSR) with the ease and speed of using PCR technology, codominant, and easily interpreted makes microsatellite the best marker in gene mapping.
Molecular Data Analysis
Data generated from the molecular analysis are often used as a basis for the classification of individuals in a population, for phylogenetic construction, parent search, identification of markers of a particular trait, taxonomic revision, and others. The dendrogram constructed in the analysis is generally based on the degree of similarity between the individuals used. However, the confidence limits for groupings generated through the dendrogram, usually not acceptable for calculations using common statistical procedures. Molecular data to be analyzed should be converted into binary data based on the presence or absence of the amplified band. If there are amplified bands scored 1, whereas if none is given a score of 0. Binary data obtained can be used to arrange the dendrogram or phylogenetic of the organism being analyzed. For parent search, genetic mapping, and identification of specific fragments of binary data are converted into genotype data, and with specific programs analyzed according to the destination.
The result of the molecular band pattern that has been converted into binary data form can be processed using Ntsys software to obtain the matrix of genetic distance and dendrogram or phylogenetic of the sample. To analyze the data according to research objectives, there is now very much software that can be used to process molecular data. For example, Genotype data composed of binary data can be used to analyze the relationship of the parent to the filial, the linkage map, and the correlation between loci with locus and locus with morphological properties. Tracking the elders using Cervus, cross-based depression in molecular markers using POPGENE, making a linkage map using MAPMAKER software with LOD 3 values, and the correlation between loci with locus and between the locus and morphological properties using statistical software such as Minitab and SPSS.
The Role and Function of Genetic Research in the Field of Aquaculture
Observation of aquaculture biota performance
Variation of phenotype as an indicator of character performance: One of the factors that determine the productivity of aquaculture is the genetic potential possessed by the cultivated population. Populations that have good genetic potential and are supported by appropriate cultivation environments, will produce better productivity compared to individuals or populations with poor genetic potential. Therefore, the selection of individuals or populations with good genetic potential needs to be done in order to improve the productivity of an aquaculture activity. One of the important parameters to be considered is the genetic variability coefficient of the population. The genetic variability coefficient is the index value that indicates the magnitude of the genetic diversity of a population. In a population with high genetic diversity, the opportunity to find individuals who have advantages over certain traits.
The performance or appearance (expression) of a character or the nature of an organism has a unique or specific or specialorder pattern. By seeing, knowing, recognizing and analyzing these unique or specific patterns of order, the breeding, selection, management, and conservation of those organisms can be done more easily, both quickly and effectively and efficiently. Performance characteristics or phenotype characteristics can be qualitative and quantitative. Qualitative qualities or traits are categorized or described traits, whereas quantitative traits or characteristics are measured traits rather than descriptive. These characteristics do not separate individuals over different phenotypic categories, such as black or albino. In fact, every character or quantitative trait shows a continuous distribution or distribution in a population. The difference between the individual phenotypes is a matter of degree or degree of difference rather than its kind.
Since the quantitative phenotype shows continuous characters/variations, the only way to learn it is to analyze the variants or variations that exist in one population and sort into the component parts. The variant or variety of phenotypes analyzed for a quantitative feature/character is the sum or combination of genetic or genotype variants, variants or various environments as well as the interactions between genetic variants and the environment. A genetic variant or variety is a combination or a resultance of an additive genetic variant, a variant or a dominant genetic variant and an epistatic genetic variant. By analyzing the qualitative and/or quantitative phenotypes, it is desirable to find patterns of order in a performance of undiscovered quantitative traits. In one population, the quantitative phenotypes are described by the central tendency as well as the distribution or distribution around the tendency. For example, mean, variance, standard deviation, co-variant, and range.
The inheritance of the quantitative nature/character is very complicated, unlike the qualitative phenotype that can be controlled by one or a pair of single genes. Quantitative phenotypes are controlled by 20 or 50 or even more than 100 genes. Even the number of genes that control the quantitative phenotype is sometimes unknown and the mode of gene expression is unknown. Whereas the inheritance of quantitative phenotypes, such as egg production, body size, body weight, growth and graduation of life, is of great importance in fish breeding efforts, both consumption fish and bait fish. This is inversely proportional to the breeding of ornamental fish that prioritizes the qualitative phenotype such as the color and shape of the fish body.
RNA and DNA ratio as an indicator of growth and nutritional conditions
The expression of RNA and DNA content in individual cells can serve as an indicator of the growth rate of the individual, this is because of seara the cytogenetic theory of the main RNA function as a protein-forming in cells that changes with cell division while the amount and size of DNA in an individual is always constant. Furthermore, the RNA / DNA ratio can be used as an indicator to describe the nutritional quality conditions consumed by aquaculture biota. Thus, RNA / DNA ratio analysis can be used to assess and evaluate the quality of aquaculture biota.
Estimating heritability as an indicator of productivity
Heritability is the proportion of population phenotypic variation caused by genetic factors. The concept of heritability is intended to assess a quantitative character in relation to the relative contribution of genetic and environmental factors to a particular trait. For example, how far the effects of genes affect habits, eg drinking alcohol, this will be useful in making social policy but this needs to be interpreted with great caution because of the high possibility of abuse misinterpretation. To assess a heritability, the first step to take is to quantify the phenotypic variation of a property, then proceed with the variance into various sources of the cause.
The superior genetic properties utilized in a cultivation business need to be controlled and maintained and maintained its genetic quality. Efforts to control and maintain superior genetic quality can be done through breeding activities. One of the things to note is knowledge of the value of heritability. Heritability shows how much a genetic factor a population affects its offspring compared to environmental factors. The greater the value of heritability in the superior traits of a population, the greater it is to obtain offspring with superior properties such as their parents.
It should be emphasized that the heritability of a trait applies only to a particular population living in a specific environment. A genetically distinct population living in an identical environment is likely to have different heritabilities for the same trait. Similarly, the same population is likely to exhibit different heritabilities for the same trait when measured in different environments, since a given genotype does not always respond to different environments in the same way. No single even genotype has superior adaptability in all environments. This is why natural selection tends to cause genetically distinct populations within a species, a population adapting typically to local conditions and not generally adapting to all environments in which it is found.
The question that always arises in quantitative traits is the question of how far a trait is genetically controlled and how far the portion of the control is by the environment. Thus, the repetitive question is how the balance of natural (genetic) versus nurture or genes versus environment influence. This is the focus of the discussion of quantitative genetics. How much variation a character (phenotype) observed is caused by genetic variation and environmental variation. To measure a phenotype variation and sort it into genetic variations and environmental variations, statistical methods are used. Example of the importance of Heritability: Selection of papaya endurance character against anthracnose can be done by knowing the character related to resistance to anthracnose, looking for genetic marker with correlation test between character (nature), heritability prediction and gene action that play role in controlling anthracnose resistance in papaya.
The heritability estimation is useful to know how big a character can be inherited. heritability is the ratio between the magnitude of the genotype and the total number of phenotypes of a character. The broad heritability value for the character of the total solid is soluble (high), the hardness of the fruit (medium) and the anthracnose resistance is high. So, these three characters have great opportunities can be genetically derived. Widely heritability estimates (h2) for endurance characters, total soluble solids and fruit hardness are high and the narrow sense heritability (h2) is moderate for the character of resistance and total dissolved solids but is low for fruit hardness.
Apart from the benefits, the heritability parameters have various limitations that are usually ignored, resulting in misuse. Hence the concept of heritability is often referred to as a parameter most often misunderstood and misused in the genetic field. Below are some of the qualifications and limitations of the heritability concept to consider:
a. heritability does not imply that a trait is genetically controlled. What is measurable from heritability parameters is the proportion of phenotypic variance among individuals in a population because of their genotype difference. This is often misunderstood and regarded as a genetically controlled trait. A gene often affects the development of a trait, so it can be said that the character is genetic. However, the difference in the phenotype between individuals in a population measured in heritability parameters is by no means genetic;
b. heritability is measured by the value of variance that can only be calculated for an individual group, so heritability is a characteristic of a population rather than an individual character;
c. heritability is not fixed for a character. There is no particular value that is common to a character. The heritability value of a character depends very much on the genotype and the environment in which the genotype grows;
d. even if there are two populations where each population has a high heritability value for very different characters, it cannot be said that the two populations are genetically different; and
e. a character possessed by individuals in a family does not always have high heritability value. Such characters are called familial trait (familial trait).
Family characters can be caused by the same genes or by the same environment, so familiality is not synonymous with heritability.
Selection and breeding program of aquaculture biota
Selection program: Selection is a program by which individuals or families are selected, in an attempt to change the average population in the next generation or, in other words, the selection is to choose individuals to be potential elders of the next generation. Selection is usually based on a minimum level of the performance index. Fish or individuals that exceed the minimum performance index will be selected as the parent stock while individuals under the minimal index or performance will be discarded or ignored. This means that the ratio or indexes measured or calculated are the performance values of individuals in a population.
In a program of character selection, quantitative traits assessed or analyzed are what genetic variants are responsible for producing such characteristics or quantitative traits that must be transmitted from parent to child in a predictable and reliable way. That is, genetic variant predictions are produced in the form of ratios or indices among the emerging quantitative character traits. For example, to predict inheritance properties such as:
a) The amount of egg production (fecundity) of a fish, then one of the quantitative traits/characteristics measured is the performance or index of gonad size and egg size. Therefore, what genetic traits that produce or cause or control the development of gonad size and fish eggs need to be measured or analyzed;
b) Growth rate, then one of the quantitative characters seen or measured is the performance or index of the size of the stomach, the length and diameter of the intestine, the size of the mouth of the mouth, etc., because to cause or produce these traits there are various genes control or manage it;
c) Life graduation or sensitivity to environmental conditions or homeostatic abilities, then one of the quantitative properties measured is the performance of the heart and other respiratory devices, because there is a genetic variation that produces or controls it; and
d) The ability or efficiency of food conversion, the predictable quantitative trait is the digestive apparatus or channels involved in this digestion process, and all these are controlled by a number of even the many different genes involved.
This means the variety or variant of the size of the stomach, gonads, heart, intestine, and others is an expression of the genetic variation that controls and produces it. Of the three variants or genetic variants of the additive variant, the dominant variant and the epistatic variant, only variants or additive variants can be transmitted in predictable and reliable ways, since the variety of additives is the function of allele, while the dominant variety is a genotype function that is the interaction between an inter alella at each locus. So that additive diversity is sometimes regarded as a determinant factor in the breeding values (breeding value) of an individual. This means that the value of performance or index-index used is a variant width (range) is considered as a variety of additives. Whether the minimum or maximum spreads that will be used as a benchmark depend on the objectives to be achieved, such as whether for ornamental fish or consumption fish, whereas the mean or even mode is the frequency of the most frequent or frequent traits that can be used to predict or predict the genetic variants of the individual.
The individual deviations seen from the performance of cardiac indexes, stomach, meat, bone, body weight, body height, etc. can be used as additive variants which are further compared or converted to the ideal size/character / healthy can be used as parameter for selection program which then sought genetic distance and cluster. In addition, the performance of the indexes for selection should be attempted on quantitative traits that can be measured at the time of the fish or the individual’s life and not when it is switched off. For example, measurement of fin indexes, eye diameter, head length, body size, body weight, and others.
Breeding program: Breeding is the application of biology, especially genetics, in the field of livestock to improve production or quality. This science is relatively new and is born as the implication of the development of human understanding of the principles of inheritance of genetic traits. In general, this science seeks to explain and apply genetic principles (with the help of other biological branches) in breeding activities. In practice, livestock breeding applies genetics, statistics, and biometrics as well as livestock reproduction, with the aim of improving the genetic quality of livestock, so as to increase production or provide added value in its implementation. From the science of genetics, related to the aspect of decreasing the nature of the elder to his offspring. Included in this are Mendel’s legal concepts. Statistics and biometrics play a role in measuring the diversity of traits and their distribution, the relationship between two or more traits, as well as analyzes for estimating genetic parameters. Reproduction is associated with aspects of fertility, fortune, breeding distance and birth.
Breeding activity is more a combination of science and art that humans have done thousands of years ago, for example: Indian maize selection and conservation in Mexico from teosinte, silkworm breeding in mainland China that produces long silk fibers, purifying various dog breeds through selection against a wolf, a horse’s cross with a mule that produces a mule, or a duckling cross with a manila duck that produces a dog. Classic breeding based on quantitative genetics is the most important approach in producing superior plant material. Some well-known strategies in breeding play a role in identifying quantitative characters using quantitative trait loci (QTL). An example of the importance of QTL is to identify the quantitative character of endocrine resistance in corn using RFLP markers. Where the identification of QTL characteristic of endurance sickness with initial help is an early breeding activity utilizing molecular marks as a tool for selection of marker assisted selection (MAS). Another example of the inheritance of quantitative characters is the inheritance test by Mendel.
Selection is a program by which individuals or groups are selected, in an attempt to change the average population in the next generation or, in other words, the selection is to choose individuals to serve as future elders of the next generation. Selection is usually based on a minimum level of performance or index. Fish or individuals that exceed the minimum performance or index will be selected as the parent stock while the individual under the index or minimum performance will be discarded or ignored. This means that the ratio or index of the index measured or calculated is the value of the performance of individuals in a population. Selection is done in the hope that the selected individual or prospective elder will inherit a population with mean and range or range similar to the value contained in the selected individual compared with the mean and the distribution range or originally. This means that the mean can be regarded as the largest frequency (mode) or range that is used as a benchmark inheritance of a genetic character.
In a program of quantitative character selection that is assessed or analyzed is what genetic variant is responsible for producing such character or quantitative traits that must be transmitted from parent to child in a predictable and reliable way. That is, genetic variant predictions are produced in the form of ratios or indices among the emerging quantitative characters [9]. For example, to predict the inheritance of traits such as the number of egg production, the rate of growth, the graduation of life and the ability to convert food. In relation to the amount of egg production (fecundity) of a fish, one of the quantitative characteristics to be measured is the performance or index of gonad size and egg size. Due to what genetic traits that produce or cause or control the development of the size of the gonads and fish eggs. Another thing is different from the rate of growth, then one of the quantitative characters that is seen or measured is the performance or index of the size of the stomach, the length and diameter of the intestine, the size of the mouth of the mouth, etc., because to cause or produce such traits there is a variety gene that control or regulate it. For a live graduation selection process or sensitive to environmental conditions or the homeostatic abilities of a cultivated biota, one of the quantitative traits measured is the performance of the heart and other respiratory devices since there is a genetic variation that produces or controls it. While the character of the food conversion ability, the predicted quantitative trait is the digestive tools or channels involved in this digestive process, and all of this is controlled by a number of even the many different genes involved. This means the variety or variant of the size of the stomach, gonads, heart, intestine, and others is an expression of the genetic variation that controls and produces it. From the three variants or genetic variants of the additive variant, the dominant variant and the epistatic variant, only variants or additives can be transmitted in predictable and reliable ways, since the variety of additives is the function of the allele, while the dominant variety is a genotype function that is the interaction between an inter allele at each locus. So this variety of additives is sometimes regarded as a determinant factor in the breeding values (values breeding) of an individual. This means that the value of performance or index-index used is a variant width (range) which is considered as an additive variant. Whether the minimum or maximum distribution that will be used as a benchmark depends very much on the objectives to be achieved eg. whether, for ornamental fish or consumption fish, the mean value or even mode is the most frequent or frequent character trait can be used to predict or predict the genetic variants of the individual.
The individual deviations seen from the performance of cardiac indexes, stomach, meat, bone, body weight, body height, etc. can be used as additive variants which are further compared or converted to the size of the ideal or characteristic of a large and healthy individual can be used as a parameter for selection program. Next look for the genetic distance and its cluster. As an example of the results displayed in the dendrogram, we can quickly sort through individuals that are the same or similar to other individuals based on the performance of the index is measured. In addition, it can be read how many degrees of similarity and differences in performance of phenotypic characters among individuals so that it can be considered in controlling or selecting individuals. Whether to continue or stop maintenance so as to save time, energy and production costs incurred. Performance data indexes and criteria made can also be used to assess the performance index of the individual price when it is offered to the market so that the selling price may be different from the selling price of other individuals who have lower performance levels.
The value of the character performance index can also be used as a guide in assessing the existence or status of the population of this species in nature whether it still reaches the standard value of the index or not. If an individual in a cultivated container exhibits a value greater than the value of this index, it may be assumed that the individual is superior and reliable, whereas if the value of the index is smaller, it can be assumed that the individual has already degraded or decreased performance so it needs anticipative steps in its management.
Engineering of aquaculture biota: Genetic engineering or genetic engineering is essentially a set of techniques performed to manipulate genetic components, ie genomic DNA or genes that can be performed in a single cell or organism, even from one organism to another organism of a different kind. In an effort to do genetic engineering, scientists use recombinant DNA technology. While organisms that are manipulated using recombinant DNA techniques are called genetically modified organisms (GMOs) that have superior properties when compared to their original organisms. Along with the advances in molecular biology today it allows scientists to take DNA from a species because DNA is easily extracted from cells. Then a molecular construction was constructed in the laboratory. DNA that has undergone molecular arrangement is called recombinant DNA while the gene isolated by the method is called a cloned gene.
Since the discovery of the structure of DNA by Watson and Crick, then began to develop genetic engineering technology in the 1970s with the aim to help create useful new products and organisms. History proves that genetic engineering constantly undergoes development and refinement of previous methods. Initially used conservative techniques pioneered by Gregor Mendel in the process of cross-breeding (breeding) to get superior seeds that are hybrid. This process takes a long time and has the disadvantage, that is emerging properties that are not cool from the plant or animal parent. Until finally was born modern genetic engineering using recombinant DNA technology. Recombination is performed in vitro (outside the cells of the organism), so it is possible to modify specific genes and transfer them between different organisms such as bacteria, plants, and animals or can cloning only one desired gene in quick time. Since the onset of the development of genetic engineering, several techniques have been continuously improved and improved in order to lead to more advanced recombinant DNA technology. The techniques that have been developed include polyploidization, androgenesis and gynogenesis, cloning, chimeras and transgenic.
Some of the steps involved in genetic engineering or recombinant DNA technology are as follows: DNA isolation containing target genes or genes of interest (GOI), isolation of bacterial DNA plasmids to be used as vectors, manipulation of DNA sequences through DNA insertion into vectors include: DNA cutting using endonuclease restriction enzymes and grafting into vectors using DNA ligase, transformation to host microorganism cells, cloning cells (and foreign genes), identifying host cells containing recombinant DNA as desired, and storage of cloned genes in DNA libraries.
Genetic engineering has penetrated in many areas, not least the field of fisheries that produce superior quality fish, for example among others: zebrafish that are usually silver with purplish black stripes, after inserted with the jellyfish color gene injected into zebrafish can then red or green from his body. The trigger gene from the jellyfish will activate the light beam on the fish when the fish is in an environment containing certain pollutant material. Transgenic carp fish with growth reaches three times its normal size because it has a gene from growth hormone salmon (rainbow trout) that is transferred directly into carp fish eggs. Similarly, other studies have produced similar results, such as in snapper (red sea bream) and Atlantic salmon which are also equally inserted by OPAFPcsGH gene growth hormone. Goldfish inserted with ocean pout antifreeze protein gene is expected to increase tolerance to cold weather. Transgenic medaka fish capable of detecting mutations (especially those caused by pollutants) is very beneficial to the life of other aquatic animals and in the field of human health. The fish is after inserted with a mutagenic mutant vector, then the DNA vector is removed and inserted into the indicator bacterium that can calculate the mutant gene. Transgenic fish become durable and not quickly rot in storage after transplanted tomato genes. But it could be otherwise if the application is intended for the world of agriculture, the fish genes that live in cold regions can be moved into the tomato to reduce the damage caused by freezing.
Induced breeding technique: Injecting is the act of entering the stimulating hormone into the female parent body. The commonly used stimulant hormone is ovaprim. Way, catch the mature female parent gonad; suction 0.6ml ovaprim for each kilogram of mains; injections of the parent’s back; parent input that has been injected into another tub and leave for 10- 12 hours. Injections can also with a solution of the goldfish hypopisa gland. Way, catch the mature female parent gonad; prepare 2kg of goldfish size 0.5kg for each female parent; cut the goldfish vertically right behind the gills tutu; cut the head horizontally just below the eye; dispose of parts of the brain; take hypopisa glands; enter the pituitary gland into the crushing glass and crush it; input 1cc aquabides and mix well; suction hypopisa solution; injections into the female parent’s back; input the parent that has been injected into another tub and leave for 10-12 hours.
Sperm picking is done half an hour before egg exposure. The trick, catch one male mature male; wipe to dry; parent body wrap with a small towel; massage into the genitalia; accommodating sperm into a plastic bowl or glass cup; mix 200cc Sodium Chloride (physiological or infusion solution); stir until homogeneous. Note: Sperm expenditure is done by two people. One person holding the head and massaging and another holding a plastic tail and bowl. Keep the sperm out of the water. Egg discharge was done after 10-12 hours after injection, but 9 hours before checking. How to remove eggs: prepare 3 pieces of plastic basin, a bottle of Sodium chloride (inpus), a chicken feather, a duster and a tissue; catch the parent with the net screw; dry the mother body with a small towel or a towel; wrap the parent with a towel and leave the egg hole open; grasp the head by one person and grasp the tail by the other; massage the abdomen toward the hole by the head holder; eggs in a plastic basin; mix the sperm solution into the egg; mix well with chicken feathers; add sodium chloride and mix thoroughly; dispose of the liquid to clean the eggs from the blood; eggs ready.
Technical induced spawning: In spawning induce spawning, eggs and sperm are not excreted, but the mother and the female are allowed to spawn on their own. Spawning is done on the tub wall. The trick, prepare ready tub wall length 4m, width 3m and height 1m; clean the mud and other dirt; dry for 3 - 4 days; water content as high as 80cm; attach hapa to the same size as the tub; female injection at 6am (see dose of injection); syringe back to the parent at 12:00 and feed into the spawning tub; injection of male parent at 12.00 and mix with the female parent; drain the water even more; let it spawn. Note: Spawning usually starts at 24.00 and ends in the morning.
Hatching grass carp fish eggs done in the aquarium. How: prepare 20 pieces of aquarium size 60cm long, 40cm wide and 40cm high; dry for 2 days; water content as high as 30cm; install four aeration points for each aquarium and turn on during hatching; spread the stock evenly to the bottom surface of the aquarium; 2-3 days then remove some of the water and add new water until it reaches its original height. Eggs will hatch in 2-3 days. Separating grass carp fish is done in a pool of land. How: prepare pool size 500m2; dry for 4-5 days; fix all parts of it; make channel of kemalir with width 40cm and height 10cm; level the ground; scatter 5-7 bags of chicken or quail dung; water content as high as 40cm and soak for 5 days (water not drained); stocking 50,000 larvae in the morning; after 2 days, give 1-2kg of pelleted flour or pellet that has been soaked daily; seed harvesting after 3 weeks.
The second nursery was also done in a pool of soil. How: prepare pool size 500m2; dry 4-5 days; fix all parts of it; make channel of kemalir with width 40cm and height 10cm; level the ground; scatter 5-7 bags of chicken or quail dung; water content as high as 40cm and soak for 5 days (water not drained); stocking 40,000 seedlings from seedling I (already selected); give 2-4kg of pelleted pellets or pellets daily; harvest the seeds done after a month old. The third nursery was done in a pool of soil. How: prepare pool size 500m2; dry 4-5 days; fix all parts of it; make its kemalir; level the ground; scatter 2 bags of chicken or quail dung; water content as high as 40cm and soak for 5 days (water not drained); stocking 30,000 heads from nursery II (selected); give 4-6kg of pellets; harvest the seeds done a month later. Enlargement of grass carp fish is done in the ground pool. How: prepare a pond size 500m2; fix all parts of it; spread 6-8 bags of chicken or quail dung; water content as high as 40- 60cm and soak for 5 days; input of 10,000 seeds selected from nursery III; feed 3 percent daily, 3kg at the start of maintenance and increase steadily in accordance with the weight of the fish; stream continuously; do the harvest after 2 months. A pond can produce fish consumption size of 125 grams as much as 400- 500kg.
Sex reversal technique: Sex reversal technology is a genital transfer technique from female to male, or vice versa, through hormonal administration and immersion techniques. If the androgen hormones are given, the fish are directed to the male sex. But if given the hormone estrogen, sex is directed to a female. So, if the cultivators want to produce bullish, then the sex reversal process applied here uses the androgen hormones. The androgen hormone used is 17-a Methyltestosterone (C20H30O2). The white, powder-shaped hormone, produced by Sigma Chemical Co., Ltd., USA, but can be bought in chemicals stores, especially major cities in Indonesia. The amount of material required 20mg/liter of solution of fish egg immersion. Each 300 fish eggs require 0.2 liters of solution. How to make a soaking solution that dissolves 10mg hormone Methyltestosterone in 0.5ml 70% alcohol, then diluted with distillate as much as 495ml. This technology is used to get the super male parent (YY), which then produces children of fish with the sex of all males. This technology is specific, so its application must be appropriate. Especially the type and dose of hormones, the duration of immersion, and the time to begin immersion. If the dose is less, then the sex of the fish will not change. But if the dose is excessive, it can cause the death of these fish. Even if not dead, the offspring tend to be sterile.
Gynogenesis technique: The purity of the carp mother must be returned. One way that can be done to restore the purity is to do inbreeding. But this method takes more than six generations. One generation takes 2 years, the time it takes to get the mother. So, this way takes 12 years. To shorten the purification period can be done by gynogenesis. This method can change from 6 generations to 2 generations, pure strains can be obtained in the second generation. The success of this method depends on the thoroughness of the treatment and fertility of female gynogenesis mentions gynogenesis is the formation of zygote 2n (diploid) without the genetic role of male gametes. So male gametes only function physically, so the process is only a parthenogenetic development of females (eggs). For that sperm irradiated. Radiation in gynogenesis aims to damage the spermatozoa chromosome so that at the time of conception does not function genetically. Nagy et al. mentions spawning by way of gynogenesis will produce all male. Gynogenesis is a rare sexual reproduction in fertilization because the sperm nuclei that enter the egg in a state of inactivity so that the development of eggs is controlled by the female genetic only. Therefore, the descendants are replicas of the female parent both morphologically and in their genetic makeup [10]. Artificial gynogenesis is performed through several treatments at the stage of conception and early embryonic development. This treatment aims to
1) To make the male genetic material inactive
2) To seek the occurrence of diploidization so that eggs can become a zygote.
Genetic material in spermatozoa is made inactive by gammaray radiation, X-rays and ultraviolet light [10]. Ultraviolet light is widely used because it is cheap.
The procedure of gynogenesis experiment: Eggs originated from broodstock mains. In order to ovulate, the parent is injected with ovaprim or hypophysis extract gland. Sperm is taken from tawes fish as much as 1ml, then diluted 100 times with a salt solution (Sodium Chloride 0.9%). After diluted in radiation with ultraviolet light for 10 minutes. Eggs and sperm mixed, resulting in fertilization. After fertilization is dissolved in plastic sieves and soaked in water with a temperature of 25 °C. After 2 minutes of fertilization in the heat shock (shock) at 40 °C for 1.5-2 minutes. To remove the stickiness of eggs given tannin solution, after that incubated at a temperature of 28 °C until hatching. The gynogenesis procedure scheme follows.
Androgenesis technique: The success of aquaculture, especially at the stage of enlargement one of them is determined by the quality of the seed. Because the seeds can live well, grow quickly, and resistant to environmental changes and disease attacks. But goldfish seeds are good quality, hard to find in Indonesia. Because the quality of the parent has dropped considerably compared to twenty years ago.
Therefore, genetics in fish should now be restored. One way of genetic improvement is by purifying the mother. One way that can be done is to do in-breeding. But this method takes more than six generations. One generation takes 2 years, the time it takes to get the mother. So, this way takes 12 years. The practical way is to pass gynogenesis. In this way, the parent purification time can be shortened to six years. Another practical way is with androgenesis, a technology that utilizes the genetic properties of fish using biotechnology principles. This technique provides the possibility to speed up the purification time in fish selection. Androgenesis can be done by manipulating some fertilization process that is making the female genetic material gametes become inactive and seek to happen diploidization.
Female genetic material gametes can be made inactive with gamma-ray radiation, x-rays or ultraviolet light [10]. Today ultraviolet light is more widely used because it is more practical and safer. Ultrasound radiation can cause chromosomal damage. Based on the research of androgenesis by Arifin, the result that radiation by using two TUV 15 wat lamps 30cm away from the egg for 3-5 minutes has been able to non-act female gamete material. Surveillance is done to maintain embryo diploidy at an early stage of its development. Diploidization can be done by inhibiting the cleavage of mitosis I. The high degree of homozygosity can be achieved by surprise at the cleavage of mitosis I because the mitotic division of the resulting pair of chromosomes is identical from the paternal haploid genome that divides into two. Without the process of embryo diploidization resulting in the fertilization of non-active eggs will be haploid characterized abnormal.
The types of surprises that can be done include temperature shocks (heat and cold), shock, shock by using chemicals and electric shock. Temperature shock is one of the many methods to do because it is easy to apply. Arai and Wilkins explained that the use of temperature shocks was easier than the shock pressure. Purdon and Lincoln suggest that heat shock has been commonly done to duplicate a set of chromosomes. In the study of androgenesis of goldfish conducted by Eddy, it was found that the time of heat shock made 40 minutes after fertilization at the best 40 °C temperature was two minutes. Research on goldfish gynogenesis showed the diploid homozygous seed produced highest by heat shock 36-37 minutes after conception. Sumantadinata states that generally the initial time of hot shock suppressing during cleavage mitosis I in gynogenesis is 40 can be done for 1.5-2.0 minutes.
The study of gynogenesis of goldfish by male parent tawes fish successfully produced gynogenetic seeds, with heat shock at 40 °C after 40 min incubation. According to Sularto et al., the highest gynogenetic production of carps fish obtained by administering a heat shock for one minute at 40 minutes after buffering. According to Sumantadinata, androgenesis is the process of embryo formation of male gametes without the female genetic contribution of females. This process of reproduction is not common, so in androgenesis is done an artificial process that disables the genetic material contained in the egg by irradiating the egg. Due to the treatment without the role of females and haploid. Haploid individuals have abnormal features such as crooked back and tail, imperfect eyes or mouth, small body size, abnormal circulatory system and inability to perform swimming and feeding activities [10]. To keep this embryo alive according to Nagy et al. needs to be carried out in the early stages of egg development. In androgenetic conducted by Arifin in goldfish managed to obtain 89.4 percent of androgenetic diploid seeds, while Eddy obtained 89.05 goldfish androgenetic seeds. Shceere et al. and Thorgaard et al. who experimented with androgenesis of rainbow fish resulted in a fish survival rate of 6.8 percent and 0.8 percent after 59 days, respectively [11-50].
Conclusion
Thus, a study or review of the role and function of genetic analysis in the field of aquaculture. From the above discussions or studies, it can be concluded that: a true and profound understanding of the concept of genetics can provide a systematic and deep pattern of thought and analysis to be applied in the field of aquaculture; and application of genetic analysis that has been applied in the field of aquaculture, among others: observation of biota character performance, selection and breeding program and engineering of aquaculture biota.
References
- Nasir M (2001) Plant Genetic Diversity. In: Makmur A (Edt.), Introduction to Plant Breeding. Directorate General of Higher Education. Ministry of National Education, Jakarta, Indonesia.
- Alif, Muhammad Dzikri (2008) Pattern of Inheritance of Some Qualitative and Quantitative Characters in Chili (Capsicum annuum L.). Bogor Agricultural Institute, Bogor, Indonesia.
- Laimeheriwa MB (2017) Phenetic Relationship Study of Gold Ring Cowry, Cypraea annulus (Gastropods: Cypraeidae) in Mollucas Islands Based on Shell Morphological. Fish Aqua J 8: 215.
- Raymond M, Rousset F (1995) GENEPOP (version 1.2.): Population Genetic Softwares for Exact Test and Ecumenicism. J Hered 86(3): 248- 249.
- Allendorf FW, Utler FM (1979) Population Genetic. In: WS Hoar, Randall DJ, Bred JR (Eds.), Fish Physiology Vol. VII. Academic Pres, New York, San Francisco, USA.
- Fajarallah A (1999) Collection of HWWDNA of Indonesia herpetofauna. Proceeding of One Daya Seminar on Life Science. Center for Life Science Study IPB.
- Higgins D (1997) Clustal V Multiple Sequence Alignments. European Molecular Biology Laboratory Germany.
- Kumar S, Tamura K, Jakobsen IB, Nei M (2001) Molecular Evolution Genetics Analysis (MEGA) Version 2.0. Oxford University Press. New York, USA Bioinformatics 17(12): 1244-1245.
- Laimeheriwa BM (2013) Analysis of the character of the quantitative phenotype in fish breeding programs. Guide to the Genetics and Breeding. Faculty of Fisheries and Marine Sciences Pattimura University, Ambon, Indonesia, pp. 25.
- Purdom CE (1993) Genetics and Fish Breeding. Chapman & Hall, London, p. 277.
- Allard RW (1951) Principles of Plant Breeding. Wiley, New York, USA.
- Aloy, Alexander B, Benjamin M, Vallejo, Marie Antonette Juinio-Menez (2011) Increased plastic litter cover affects the foraging activity of the sandy intertidal gastropod Nassarius pullus. University of the Philippines, Philippines, Marine Pollution Bulletin 62(8): 1772-1779.
- Anthony JF Griffiths, Jeffrey H Miller, David T Suzuki, Richard C Lewontin, William M Gelbart (2000) An Introduction to Genetic Analysis. In: WH Freeman (Edt.), Genes as determinants of the inherent properties of species. The University of British Columbia, University of California, Harvard University (7th edn).
- Ayala F, Kiger JA (1984) Modern Genetics. The Benjamin Cummings, Menlo Park, California, USA, 13(2): 87.
- Campbell, Neil A Reece, Jane B, Mitchell Lawrence (2010) Biology Volume I, (8th edn), Erlangga, Jakarta, Indonesia.
- Crowder LV (2010) Plant Genetics. Gadjah Mada University Press, Yogyakarta, Indonesia, pp. 499.
- Edelman A (1988) Eigenvalues and condition numbers of random matrices. SIAM Journal of Matrix Analysis and Applications 9(4): 543- 560.
- Elizabeth P (2007) DNA Study Forces Rethink of What It Means to Be a Gene. Science 316(5831): 1556–1557.
- Falconer DS, Mackay TFC (1996) Introduction to Quantitative Genetics. (4th edn), Addison Wesley Longman, Harlow, Essex, UK.
- Falconer DS (1981) Introduction to quantitative genetics. John Wiley and Sons.
- Falconer (1996) Introduction to quantitative genetics. Longman.
- Fisher RA (1930) The Genetical Theory of Natural Selection. Clarendon Press, Oxford, UK.
- Gordon IL (2003) Refinements to the partitioning of the inbreed genotypic variance. Heredity 91(1): 85-89.
- Hair JR (2010) Multivariate Data Analysis (7th edn), Pearson Prentice Hall, USA.
- Kaufman I, Rousseuw PJ (2008) Finding groups in data: an introduction to cluster analysis. 344.
- King RC, Stansfield WD (1985) A dictionary of genetics. (3rd edn), Oxford University Press Inc., New York, USA.
- Leimeheriwa BM (2012) Guidance for Assessment of Quantitative Phenotypes in Hatching Units. Faculty of Fisheries and Marine Sciences Pattimura University Ambon, Indonesia, pp. 28.
- Laimeheriwa BM (2016) Morphology and Meristic Characters for Numerical Taxonomy of Shell’s Gastropods. MMCSE, Pattimura University, Ambon, Indonesia, p. 103.
- Laimeheriwa BM (2017) The Construction of Model Numerical Taxonomy and Determination of Character Performance Index of Ring Snail, Cypraea annulus, LINN. 1758. Dissertation. Pattimura University, Ambon, Indonesia, p. 560.
- Lynch M, Walsh B (1998) Genetics and Analysis of Quantitative Traits. Sinauer, Sunderland, Massachusetts, USA.
- Mather (1971) Biometrical genetics. Chapman and Hall, USA.
- McGlade JM, dan Boulding E (1986) The truss: A geometric and statistical approach to the analysis of form in fishes. Can Tech Rep Fish Aquacult Sci p. 147.
- Mendel Gregor (1865) Experiments in plant hybridization. Verhandlungen naturforschender Verain iv. für das Jahr Abhandlungen, pp. 3–47.
- Nei M (1972) Genetic Distance Between Population. American Naturalis 106(949): 283-284.
- Pai AC (1985) The Basics of Genetics. Erlangga, Jakarta, Indonesia.
- Pearson H (2006) Genetics: what is a gene?. Nature 441(7092): 398– 401.
- Peter J Bowler (1989) The Mendelian Revolution: The Emergency of Hereditarian Concepts in Modern Science and Society Johns Hopkins University Press, Baltimore, USA 26(4): 207.
- Roff DA (1997) Evolutionary Quantitative Genetics. Chapman & Hall, New York, USA.
- Ryman N, Utter F (1987) Population genetics and fishery management. University of Washington Press, Seattle, USA, p. 420.
- Seykora, Tony (2011) Animal Science 3221 Animal Breeding. Tech. University of Minnesota, Minneapolis, USA.
- Stansfield WD (1989) Schum’s outline of genetics. McGraw-Hill, New York, USA.
- Strickberger MW (1985) Genetics. Macmillan Publisher Co. Inc, New York, USA.
- Suryo (2010) Human Genetics, Gajah Mada University Press, Yogyakarta, Indonesia.
- Suryo H (2007) Cytogenetics. Gadjah Mada University Press, Yogyakarta, Indonesia, p. 446.
- Suryo H (2012) Genetics for Strata 1. Gadjah Mada University Press, Yogyakarta, Indonesia, p. 345.
- Tave D (1993) Genetics for fish hatchery managers. Van Nostrand Reinhold, New York, USA.
- Tave D (1995) Selective Breeding Programme for the medium-sized fish farm. FAO Fish. Tech Paper No. 352. Rome, Italy, p. 122.
- Veer B (2014) Biostatistics Basics. Karisma Publishing Goup, Jakarta, Indonesia, p. 512.
- Wright S (1951) The genetical structure of populations. Annals of Eugenics 15(4): 323-354.
- Yatim W (1991) Genetics. Tarsito, Bandung, Indonesia.






























