Utilization of M-PCR and P-AST for Diagnosis and Management of Urinary Tract Infections in Home-Based Primary Care
Annemarie Daly1, David Baunoch2*, Kelly Rehling1, Natalie Luke2, Meghan Campbell2,3, Patrick Cacdac2, Miguel Penaranda2, Michael Opel2, Shuguang Huang4, Xinhua Zhao4 and Kirk Wojno1
1 Visiting Physicians Association, 500 Kirts Blvd, Suite 200, Troy, Michigan
2 Pathnostics, Inc, 17661 Cowan, Irvine, CA
3University of Kentucky College of Medicine, Lexington, KY
4 Stat4ward, 711 Parkview Dr, Gibsonia, PA
Submission: March 13, 2020;Published: March 27, 2020
*Corresponding author: David Baunoch, Pathnostics, 17661 Cowan, Irvine, CA
How to cite this article: Annemarie D, David B, Kelly R, Natalie L, Meghan C, et al. Utilization of M-PCR and P-AST for Diagnosis and Management of Urinary Tract Infections in Home-Based Primary Care. JOJ Urology & Nephrology, 2020; 7(2): 555707. DOI: 10.19080/JOJUN.2020.07.555707
Abstract
Background/Objective: Urinary tract infections are common, especially among the elderly, and are associated with higher rates for both emergency department utilization and hospitalization. Improvements in diagnostic accuracy and timeliness may enable better outpatient care and thereby reduce emergency department utilization and hospital admission rates.
Methods: This was a retrospective study of existing data from 66,383 patients seen for possible urinary tract infection by house-call primary care providers. Patients were divided into two cohorts. One cohort of patients was treated based upon the results from Standard Urine Cultures (SUC). The other cohort was treated in accordance with results from an assay combining Multiplex-Polymerase Chain Reaction (M-PCR) and Pooled Antibiotic Susceptibility Testing (P-AST) of urine specimens. The total number of emergency department visits and hospitalizations were compared between the two cohorts.
Results: We found that the use of the combined M-PCR/P-AST was associated with a 13.7% decrease in hospital admissions and/or emergency department utilization when compared to the use of SUC testing (3.27% vs. 3.79%; p = 0.003).
Conclusions: These findings suggest that use of a combined M-PCR/P-AST assay in outpatient management of suspected urinary tract infection may improve patient outcomes and reduce emergency department and hospital utilization.
Keywords: Urinary tract infection; Cystitis; Elderly; Emergency department; Hospitalization; Home-based primary care
Abbreviations: UTI: Urinary Tract Infections; ED: Emergency Department; M-PCR: Multiplex Polymerase Chain Reaction; P-AST: Pooled Antibiotic Susceptibility Testing; SUC: Standard Urine Culture; CD: Charlson/Deyo; STDDIFF: Standardized Difference; SD: Standard Deviation; IQR: Interquartile Range; VPA: Visiting Physicians Association; CMS: Center for Medicare and Medicaid Services
Introduction
Urinary Tract Infections (UTI) are among the most common infections, accounting for 10.5 million office visits and 3 million Emergency Department (ED) visits in the U.S. annually, costing $3.5B in 2007 [1]. In 2011, 436,437 patients were admitted to the hospital for UTIs at the cost of $2.8B [2]. UTI’s are a leading cause of infection-related hospitalization events in the elderly [2,3]. Properly managing UTI’s in the community leads to reduced healthcare costs. In studies evaluating the management of complicated UTI’s, patients who had their symptoms managed in an outpatient setting had a total expense of $17,914 for all treatment associated cost. Patients who had an associated ED visit resulting in hospitalization had a total treatment associated expense of $82,153. $64,239 is averted when a single patient avoids hospitalization/ED for a UTI [4].
Since the early 1950s, the standard of care for diagnosis of UTI is SUC, along with antimicrobial susceptibility testing. The SUC methodology relies on an “Escherichia coli (E. coli)-centric” view that perceives UTIs as caused by one or two pathogens [5]. Recent findings, however, reveal that SUC misses up to 89% of all uropathogens; furthermore, up to 39% of these infections are polymicrobial [3,6,7]. Consistent with previous reports, we’ve also found that polymicrobial infections constituted over 30% of all symptomatic patients [6]. We’ve found M-PCR to be superior to SUC for the detection and identification of bacteria in the context of UTIs, along with the ability to detect and identify bacteria in polymicrobial infections. Along with M-PCR, we’ve utilized P-AST to determine antibiotic susceptibility and resistance of the organisms identified by M-PCR. Antibiotic susceptibility assays that test individual pathogens against antibiotics may miss interactions between bacteria in polymicrobial environments. The P-AST tests all pathogens within a urine specimen simultaneously against antibiotics and can be informative about changes in antibiotic response due to bacterial interactions. We recently showed that bacterial interactions in polymicrobial infections could alter antibiotic susceptibilities. These changes can significantly affect the type and dosage of antibiotics required to treat the patient [8]. Further, the use of the M-PCR/P-AST test significantly reduces turnaround time, providing results in 24 hours, with SUC taking up to 5 days to result. Thus, concurrently, the M-PCR/P-AST assay quickly detects 2 organisms or more while also providing susceptibility information in polymicrobial samples [6]. In the present retrospective study, we evaluated if M-PCR/P-AST was superior to SUC using ED utilization and hospitalization rates as proxy measures of outpatient treatment effectiveness: more effective outpatient treatment should result in lower rates for ED utilization and hospitalization.
Methods
Study design and participants
This was an observational, retrospective study that compared the number of ED visits and hospital admissions between two patient cohorts. The study population consisted of patients seen by primary care providers who attend to patients in their homes or assisted living facilities. Patients residing in the following states were included in this study: Arizona, Florida, Illinois, Indiana, Kansas, Kentucky, Michigan, Missouri, North Carolina, Ohio, Texas, Virginia, Washington, and Wisconsin, along with the District of Columbia.
All patients in this study were seen for possible UTI. Patients were divided into two time period cohorts. The SUC cohort consisted of patients who received outpatient treatment with antibiotics based upon results from SUC between March 1, 2016 and July 31, 2017. There was a washout period of 7 months (August 1, 2017 – February 28, 2018). The M-PCR/P-AST cohort included patients who received outpatient treatment with antibiotics based upon results from M-PCR testing between March 1, 2018 and July 31, 2019.
Urine culture
Urine samples were obtained from patients by either selfadministered clean catch or catheterization. Samples were collected and transported to Lab Corp (Various Locations in US) or Quest Diagnostics (Various Locations in US) for testing by culture using standard procedures.
DNA extraction and analysis
DNA extraction was performed using the King Fisher/ MagMAX™ Automated DNA Extraction instrument and the MagMAX™ DNA Multi-Sample Ultra Kit (Thermo Fisher, Carlsbad, CA). 400 μL of urine were transferred to 96-well deep-well plates, sealed, and centrifuged to concentrate the samples, and then the supernatant was removed. Enzyme Lysis Mix (220 μL/well) was added to the samples, which were then incubated for 20min at 65ºC. Proteinase K Mix (PK Mix) was added (50 μL/well) and incubated for 30min at 65ºC. Lysis buffer (125 μL/well) and DNA Binding Bead Mix (40μL/well) were added, and the samples were vortexed for a minimum of 5 min. Each 96-well plate was loaded into the KingFisher/MagMAX Automated DNA Extraction instrument, which was operated in accordance with standard operating procedures.
DNA analysis was conducted using the Guidance® UTI Test (Pathnostics, Irvine, CA), which consists of both M-PCR and P-AST. Samples were mixed with universal PCR master mix and amplified using TaqMan technology on the Life Technologies 12K Flex OpenArray System™ (Life Technologies, Carlsbad, CA). DNA samples were spotted in duplicate on 112-format OpenArray chips. Plasmids unique to each bacterial species being tested were used as positive controls. Candida tropicalis was used as an inhibition control. A data analysis tool developed by Pathnostics was used to sort data, assess the quality of data, summarize control sample data, identify positive assays, calculate concentrations, and generate draft reports. Probes and primers were used to detect the following pathogenic bacteria: Acinetobacter baumannii, Actinotignum schaalii, Aerococcus urinae, Alloscardovia omnicolens, Citrobacter freundii, Citrobacter koseri, Corynebacterium riegelii, Enterobacter aerogenes, Enterococcus faecalis, Escherichia coli, Klebsiella oxytoca, Klebsiella pneumoniae, Morganella morganii, Mycobacterium tuberculosis, Mycoplasma genitalium, Mycoplasma hominis, Pantoea agglomerans, Proteus mirabilis, Providencia stuartii, Pseudomonas aeruginosa, Serratia marcescens, Staphylococcus aureus, Streptococcus agalactiae, and Ureaplasma urealyticum. Probes and primers also were used to detect the following bacterial groups: Coagulase Negative Staphylococci (CoNS) (Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus lugdunensis, Staphylococcus saprophyticus); Viridans Group Streptococci (VGS) (Streptococcus anginosus, Streptococcus oralis, Streptococcus pasteuranus). Reporting included both the name of the organism identified and semi quantified counts of organisms. Counts were reported in cells/mL and correlated to Colony Forming Units (CFU).
Pooled–Antibiotic Susceptibility Testing
P-AST was performed by aliquoting 1mL of patient urine specimen into a 1.7mL microcentrifuge tube. After centrifugation, the supernatant was aspirated and discarded, leaving approximately 500μL of patient sample in the microcentrifuge tube. One mL of Mueller Hinton Growth Media was then aliquoted into the patient sample in the microcentrifuge tube and the tubes were incubated at 35ºC in a non-CO2 incubator for 6 hours. Mueller Hinton Agar was used as a negative control. Those samples that reached a minimum threshold of 10,000 cells/mL were then diluted by aliquoting 0.5mL of sample into a 50mL conical tube containing Mueller Hinton Growth Media. 96-well plates preloaded with antibiotics were then inoculated with diluted samples and incubated along with control plates for 12-16 hours at 35ºC in a single layer. Optical density of samples was then read on a DensiCHEK plate reader™ (BioMerieux, Marcy-l’Étoile, France).
P-AST is currently unavailable for U. urealyticum, A. schaalii, A. urinae, A. omnicolens, C. riegelii, M. hominis, and M. genitalium due to fastidious in vitro growth characteristics.
Data Collection
Patient demographic, comorbidity, and location (State) were collected for both groups of patients using the Health Catalyst (data warehouse) and are presented in Table 1. Demographic variables included age, gender and race. Additionally, the Charlson/Deyo (CD) Index Score was collected along with the number of perpatient physician visits.
ED visit and hospital admission rate for UTI were identified using the primary diagnosis code from files available through the Centers for Medicare and Medicaid Services (CMS). Study data included medical records with both ICD-9 and ICD-10 codes. ICD- 9 codes were still being used during the time period for the SUC cohort until the transition to ICD-10 coding took effect during the time period for the M-PCR/P-AST cohort. Applicable ICD-9 diagnosis codes include 5990 (Urinary tract infection), 788.1 (Dysuria), 590.10 (Acute pyelonephritis), 590.80 (Pyelonephritis), 5999 (Urinary tract disease), and 59000 (Chronic pyelonephritis). ICD-10 diagnosis codes include N39.0 (Urinary tract infection, site not specified), R30.0 (Dysuria), R35.0 (Frequency of micturition), R32 (Unspecified urinary incontinence), Z87.440 (Personal history of urinary (tract) infections). Cases were excluded if records indicated that the National Provider Identifier (NPI) did not match that of a listed investigator participating in the study, if a patient resided in hospice, or if the diagnosis code was missing.
Statistical Analysis
Patient demographic and comorbidity data are presented in Table 1. Differences between the cohort 1 (SUC) and cohort 2 (M-PCR/P-AST) cohorts were analyzed using standardized difference (STDDIFF). The STDDIFF was calculated as the difference in means (or proportions) divided by the pooled estimate of the standard deviation, with the value of 0.1 considered negligible difference and 0.2 to 0.49 considered a small difference between the two groups [9]. STDDIFF is increasingly used to compare balances in baseline covariates between study groups because unlike other statistical tests, such as Student’s t-test that produces p values; the STDDIFF is not influenced by the sample size. At the same time, it also allows for the comparison of the variables measured in different units.
We calculated the average number of ED visits and/or hospitalizations for UTI for each cohort. A generalized linear model was used to compare the number of ED visits and/or hospitalizations between the two cohorts, using the negative binomial distribution with log link to account for over-dispersion of the count data. The dependent variable was the count data for the number of ED visits and/or hospitalizations per patient per cohort, and the independent variable was the cohort. Multiple UTI events per patient could confound the number of ED visits and/ or hospitalizations. We therefore also conducted a patient-level analysis by summarizing the frequencies and the proportion of patients with any ED or hospitalization event for UTI. A logistic regression model with logit link was used for this binary outcome to assess the difference between the two groups. The dependent variable was the dichotomized variable of any hospitalization and/or ED visit per patient per cohort; the independent variable was the cohort.
Finally, we described the per-patient frequencies of hospitalization/ED visits by the two cohorts, in order to elucidate the number of patients who experienced multiple events. The age distribution by the number of ED or hospitalization and cohort is displayed in Figure 1. The analyses were performed using SAS version 9.4 (SAS Institute, Inc., Cary, NC) and StataSE 16.
Results
Overall, the retrospective chart review included a total of 66,381 patients, with 34,414 unique patients in the SUC cohort and 31,967 unique patients in the M-PCR/P-AST cohort. Table 1 presents the patient demographics, co-morbidity, and location for each group. The two cohorts were similar, as the STDDIFF for demographic variables were less than 0.20 for all variables. The mean age for the SUC cohort was 73.5 years and 72.9 years for the M-PCR/P-AST cohort. The gender breakdown was similar in the two cohorts, with 62.9% of patients being female in the SUC cohort and 62% female in the M-PCR/P-AST cohort. There was a negligible difference in the number of patients seen in Ohio, Florida, and Washington. Compared to the SUC cohort, less patients were seen in Ohio for the M-PCR/P-AST cohort (17.1% v 23%, STDDIFF = -0.148) while more patients were seen in Washington (3.4% v 1.5%, STDDIFF = -0.126) and Florida (11.7% v 8.3%, STDDIFF = 0.112). The mean CD Risk Score was 3.9, and the median was 4.0 for both cohorts. The race distribution was also similar in the two cohorts, with 73.6% of patients being white in the SUC cohort and 75.2% white in the M-PCR/P-AST cohort. The mean number of physician visits was 6.2 for the SUC cohort and 6.1 for the M-PCR/P-AST cohort, with a median of 5.0 for both cohorts.
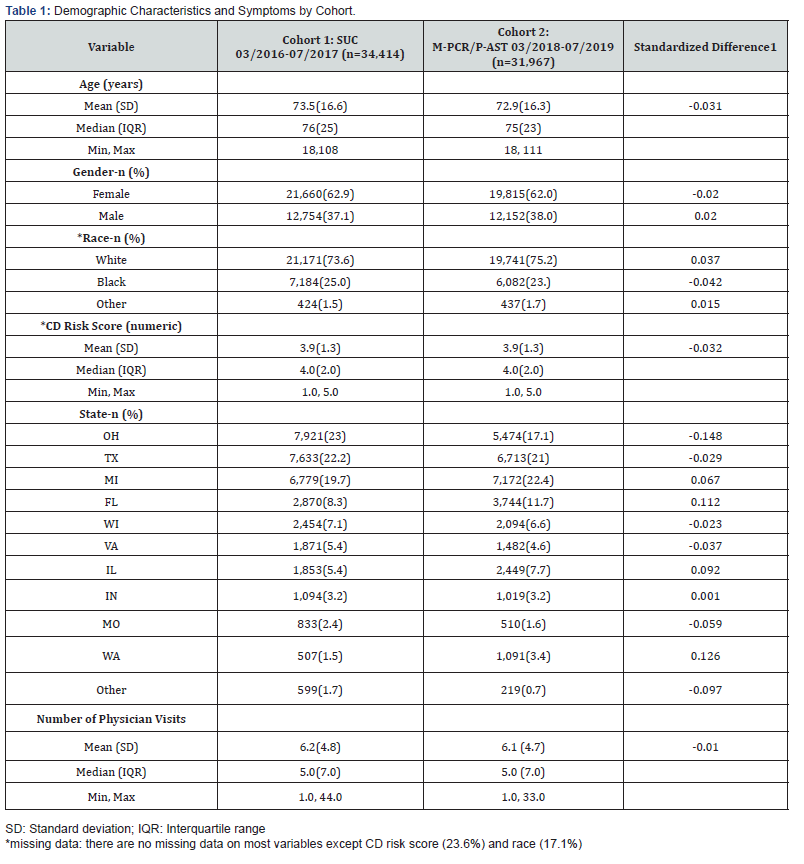
[1]Standardized difference was calculated as the difference in means (or proportion) divided by the pooled estimate of the standard deviation, with an absolute value of 0.1 considered negligible difference and the absolute value 0.2-0.49 considered as a small difference between the two groups.
Summary statistics and a comparison of ED visits and hospital admission rates between the two cohorts are presented in Table 2. Compared to patients in the SUC cohort, patients in the M-PCR/P-AST cohort visited the ED and/or were admitted to the hospital less frequently. Of the 34,414 patient cases in the SUC cohort, there were a total of 1,307 (3.79%) events of either ED visits or hospitalizations during the study period. By contrast, 1,045 (3.27%) similar events took place in the 31,967 patient cases reviewed in the M-PCR/P-AST cohort, which translated to a statistically significant (p = 0.003) difference between the two cohorts.
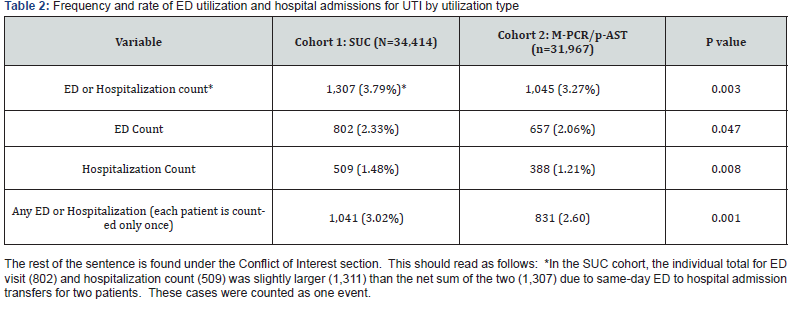
Overall, the number of ED visits decreased between the two cohorts, from 802 (2.33%) for the SUC cohort to 657 (2.06%) for the M-PCR/P-AST cohort, but this difference bordered on statistical significance (p = 0.047). The number of hospitalizations was significantly lower in the M-PCR/P-AST cohort (388; 1.21%) than in the SUC cohort (509, 1.48%; p = 0.008). Multiple hospitalizations per patient could confound the number of hospitalizations. If each patient was counted only once per UTI event, there was a statistically significant reduction in patients either utilizing the ED or being hospitalized, from 1,041 (3.02%) for the SUC cohort to 831 (2.60%, p = 0.001) for the M-PCR/PAST cohort. Table 3 presents the patient-level distribution of ED or hospitalization frequencies. One patient could experience multiple ED or hospital utilization events over the study timeperiod, with a maximum of nine events per patient. Slightly fewer patients in the M-PCR/P-AST cohort (150, 0.47%) experienced multiple events than the SUC cohort (180, 0.52%; p = 0.32). Figure 1 displays relationships between age and number of ED visits and/or hospitalizations. Overall, those patients not experiencing an ED/hospitalization event, or experiencing 1-3 events were similar in age. From the graph, we see a trend that patients in the M-PCR/P-AST cohort who experienced 4 or more visits were older than patients in the SUC cohort, yet from the results above (Table 3) we see that the ED visit and hospitalization rates for the M-PCR/P-AST cohort were smaller than the SUC cohort.


Discussion
UTIs are common in the elderly and have the potential to create significant health complications if the appropriate measures are not taken in a timely manner. UTIs are a common cause of infection-related hospitalization [1]. For this study, the population observed were elderly requiring home-based care to meet their healthcare needs. These patients often have chronic conditions and functional limitations that make it difficult or impossible to travel to a physician’s office. More efficacious treatment of UTIs in this population may lead to decreased hospitalizations/ED visits, thereby reducing healthcare costs.
In this study, we evaluate all unique patient samples collected by the Visiting Physicians Association (VPA), a multi-state homehealth provider for patients presenting with symptoms of urinary tract infection. It attempts to assess the impact transitioning testing for their patients, from SUC to M-PCR/P-AST, had on ED and hospitalization rates. VPA had moved away from SUC based on reports in the literature suggesting that SUC had a >30% falsenegative rate and missed significant numbers of uropathogens [7]. An additional motivation in internalizing this test was to reduce the turn-around associated with SUC testing to decrease empiric treatment. The study sample was divided into two cohorts, one in which UTI treatment decisions were made based upon results from SUC and antimicrobial susceptibility testing, and the other in which UTI treatment decisions were made based upon results from M-PCR/P-AST. The two cohort populations were similar in age, gender, race, CD Risk Score, location, and the number of provider visits.
We found a significant association between the use of the M-PCR/P-AST assay and a reduction in ED utilization and hospital admission rates. Specifically, ED utilization and hospital admission rates for UTI were higher in patients for whom prior outpatient treatment was guided by results from SUC than patients for whom prior outpatient treatment was guided by results from the M-PCR/ P-AST assay. This association was stronger for hospitalization rates than for ED utilization. Further, there were fewer repeated hospitalization events for patients managed based upon results from the M-PCR/P-AST assay (0.47%, 150/31,967) than for patients managed using SUC (0.52%, 180/34,414, p = 0.32).
While the percentage of patients in this population being sent to the ED or hospital is relatively small, the cost impact of the reduction is significant when considering a large patient population of 30,000. Another study examined the costs of managing a UTI. They analyzed the charges incurred over three time periods, within 30 days of the initial UTI event, ED/ Hospitalization, and during UTI follow up. Expenses connected with these 3 time periods include the cost of the physician visits, any pharmaceuticals used, and follow up visits. Hospitalized patients incurred more charges than those that non-hospitalized patients. These charges include the fees acquired while in the hospital and during the follow-up visits. In total, $65,239 more was spent on patients who experienced an ED and/or hospitalization event compared to non-hospitalized visits [4]. Thus, a 13.7% reduction in hospitalization when normalized against cohort 1 would result in a 156 patient decrease in ED/hospitalizations and/or ED utilization leads to a $10,177,284 savings. Based on the results of the study VPA instituted the test as standard of care for their patient population.
The strength of this study is the ability to examine hospitalization/ED events and the impact of testing devices in a large population. Even with an exhaustive analytical approach to the large dataset, we didn’t have the means to trace back specific hospital events to a specific physician visit and test result. Because we were not able to trace back to a specific physician visit, we were not able to identify a specific relationship between hospitalizations to a test result. The absence of a direct relationship means that we were unable to show a direct causation. Thus, we found an association between the use of the M-PCR/P-AST and reductions in both ED utilization and hospitalization rates when comparing the two large cohorts. In this study we have found an association between the use of the M-PCR/P-AST and reductions in both ED utilization and hospitalization rates when comparing two large cohorts. Future studies involving randomization are underway to investigate further the role that M-PCR/P-AST may aid in the management of UTI in the elderly population.
Conclusion
This study compared an assay that combines M-PCR detection of pathogens and P-AST with traditional urine culture in the context of UTI treatment. We found a statistically significant association between the use of M-PCR/P-AST and reductions in both ED use and hospitalizations. Greater diagnostic accuracy and shorter turn-around time provided by M-PCR/P-AST may have contributed to this finding. Additional studies are planned to assess the degree to which M-PCR/P-AST may improve clinical outcomes in elderly patients.
Conflict of Interest
Financial assistance for the study was provided for by Thermo Fisher and Pathnostics
Corrigendum to:
“Utilization of M-PCR and P-AST for Diagnosis and Management of Urinary Tract Infections in Home-Based Primary Care” by Annemarie Daly et al. (JOJ Urology & Nephrology, 2020; 7(2): 555707. DOI: 10.19080/JOJUN.2020.07.555707) Based upon Turner et al., there were $64,239, rather than $65,239, more costs involved in managing patients who required hospitalization for UTI. The additional costs resulted in $10,021,284 more being charged to the health care system when applied to a population of 30,000 patients rather than $10,177,284.
References
- Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ (2015) Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nature reviews Microbiology 13(5): 269-84.
- Simmering JE, Tang F, Cavanaugh JE, Polgreen LA, Polgreen PM (2017) The Increase in Hospitalizations for Urinary Tract Infections and the Associated Costs in the United States, 1998-2011. Open forum infectious diseases 4(1): ofw281.
- Hilt EE, McKinley K, Pearce MM, Rosenfeld AB, Zilliox MJ, M, et al. (2014) Urine Is Not Sterile: Use of Enhanced Urine Culture Techniques To Detect Resident Bacterial Flora in the Adult Female Bladder. Journal of Clinical Microbiology 52(3): 871-876.
- Turner RM, Wu B, Lawrence K, Hackett J, Karve S, et al. (2015) Assessment of Outpatient and Inpatient Antibiotic Treatment Patterns and Health Care Costs of Patients with Complicated Urinary Tract Infections. Clinical Therapeutics 37(9): 2037-2047.
- Price TK, Hilt EE, Dune TJ, Mueller ER, Wolfe AJ, et al. (2018) Urine trouble: should we think differently about UTI?. Int Urogynecol J 29(2): 205-210.
- Wojno KJ, Baunoch D, Luke N, Opel M, Korman H, et al. (2019) Multiplex PCR Based Urinary Tract Infection (UTI) Analysis Compared to Traditional Urine Culture in Identifying Significant Pathogens in Symptomatic Patients. Urology 136: 119-126.
- Price TK, Dune T, Hilt EE, Thomas-White KJ, Kliethermes S, et al. (2016) The Clinical Urine Culture: Enhanced Techniques Improve Detection of Clinically Relevant Microorganisms. Journal of Clinical Microbiology 54(5): 1216-1222.
- Vollstedt A (2020) Resistance Patterns as Detected by Urobiome Antibiotic Susceptibility Testing (U-AST) in Polymicrobial Urinary Tract Infections.
- Hardin JW, Hilbe JM (2012) Generalized Estimating Equations.






























