In Silico Analysis of NPHS1 Gene Mutations Identified in Patients with Steroid Resistant Nephrotic Syndrome
Madiha Shakoor1, Farkhanda Hafeez2, Shahida Praveen2, Faiza Noor2, Ali Amar1, Ayesha Abid1 and Shagufta Khaliq1*
1Department of Human Genetics and Molecular Biology, University of Health Sciences, Lahore, Pakistan
2Department of Human Genetics and Molecular Medicine, Sindh Institute of Urology and Transplantation, Karachi, Pakistan
3Department of Pediatric Nephrology, The Children Hospital, Lahore, Pakistan
Submission: February 08, 2017; Published: April 28, 2017
*Corresponding author: Shagufta Khaliq, Department of Human Genetics and Molecular Biology, University of Health Sciences, Lahore, Pakistan, Tel: +924299231304-9; E-mail: khaliq.shagufta@gmail.com
How to cite this article: Madiha S, Farkhanda H, Shahida P, Faiza N, Ali Amar, Aiysha A Shagufta K. In Silico Analysis of NPHS1 Gene Mutations Identified in Patients with Steroid Resistant Nephrotic Syndrome. JOJ uro & nephron. 2017; 2(5): 555599.DOI: 10.19080/JOJUN.2017.2.555599
Abstract
Introduction: Steroid Resistant Nephrotic syndrome (SRNS) is a rare autosomal recessive kidney disorder caused by increased permeability of the glomerular capillary wall for macromolecules leading to heavy proteinuria, hypoproteinemia and edema. SRNS patients do not respond to steroid therapy and have a poor prognosis with high risk of end-stage renal failure. SRNS is caused by the genetic defects in podocyte expressed proteins which do not lead to recurrence after renal transplantation. To date several mutations in podocyte genes, NPHS1&2 have been associated as an underlying genetic factor. Here we investigated the pathogenic effects of the NPHS1 gene mutations reported in Pakistani patients.
Methods: In-Silico analysis was carried out to study the effects of previously reported 7 different mutations by using software's i.e. SIFT, PROVEAN, Mutation Assessor, MSA, I-TASSER and PyMol.
Results: Pathogenicity prediction tools and MSA predicted the damaging nature of these mutations and their conservation across the species respectively. Structural modeling using the I-TASSER and subsequent visualization by PyMol software demonstrated the position and effects of these mutations on different domains of nephrone protein.
Conclusion: Analysis predicted the potential deleterious effects of these mutations and showed interruption of nephrone protein with surrounding structural proteins that might have led to the disruption of filtration barrier resulting in SRNS.
Keywords: Steroid Resistant nephrotic syndrome; Podocyte; NPHS1; Gene mutations; In silico analysis
Abbreviations:NS: Nephrotic Syndrome; CNS: Congenital Nephrotic Syndrome; SSNS: Steroid Sensitive Nephrotic Syndrome; SRNS: Steroid Resistant Nephrotic Syndrome
Introduction
Nephrotic syndrome (NS) is autosomal recessive glomerular disease caused by increased permeability of the glomerular capillary wall for macro molecules. It is characterized by heavy proteinuria followed by hypoproteinemia, hypercholesterolemia, lipiduria and edema [1,2]. Diagnostic evaluation and prognostic classification depends on disease onset and response towards therapy. Differential diagnosis of NS can be narrowed down on the bases of disease onset as congenital, infantile and childhood. Congenital nephrotic syndrome (CNS) appears in utero or during the first 3 months of life. In case of Infantile NS, the onset of the disease starts between the ages of 4 months to first year of life, whereas childhood onset NS cases are diagnosed after first year of life [3]. On the bases of patient's response towards steroid therapy, data suggested two different types of nephrotic syndromes, steroid sensitive nephrotic syndrome (SSNS) and steroid resistant nephrotic syndrome (SRNS). SSNS patients are managed by immunosuppressive therapy however; almost 20% patients with NS fail to respond steroid therapy and referred as SRNS having persistent proteinuria after 4 weeks of oral steroid therapy [4,5]. It is a life-threatening condition and renal transplantation is the only therapeutic option.
In the renal system, the glomerular filtration barrier (GFB) of kidney serves as ultra-filter for plasma [6]. Its capillary wall is composed of 3 layers of cells i.e., fenestrated endothelium, glomerular basement membrane and the podocyte. Podocytes are highly specialized cells which consist of large cell body and elongated cellular extensions called foot processes. The adjacent foot processes bridge by a slit known as slit diaphragm or filtration slit. It is a zipper-like structure of 2540nm, the approximate size of an albumin molecule [7,8]. The integrity of structural proteins, including nephrin, podocin, CD2-associated protein, P-cadherin, catenins and nephrin like 1-3 serve to preserve the normal podocyte architecture [9, 10]. Their structural disruption causes proteinuria leading to SRNS and many other glomerular diseases. Genetic factors are major cause of glomerular disease and understanding of these factors at molecular level is a valuable diagnostic tool for the selection of therapy. Genetic factors are further divided into two categories. The structural defects due to mutations do not lead to recurrence after renal transplantation because donor's kidney does not have such defects. Whereas, immunological factors like production of B and T cell-dependant inflammatory cytokine against GFB disrupt the kidney function by changing the permeability of the filtration barrier and lead to recurrence even after renal transplantation [11-13].
SRNS is genetically a heterogeneous disorder as mutation(s) in various genes are involved in the pathogenesis of disease [14] of these NPHS1 (19q13) gene is reported as one of the most common disease causing gene in patients with SRNS. The NPHS1 gene encodes nephrin protein consisting of 1241 amino acid which is an essential component of the slit diaphragm (SD). The NPHS1 knock-out mice model showed an effacement of pedicels and absence of slit diaphragm in their kidneys resulting in proteinuria and neonatal death [15-17]. NPHS1 gene mutations are most commonly reported in Finland with the incidence of 1:10,000 live births. More than 200 functionally significant nephrin mutations had also been reported in non-Finnish patients from Malta, Europe, Old Order Mennonites, North Africa and America. [18-20]. In addition, study conducted on Pakistani SRNS patients reported 7 mutations in NPHS1 gene of which 6 were novel [21]. In view of such a wide spectrum of disease associated NPHS1 gene mutations, this study was carried out to predict the pathogenicity and the effect of these mutations on nephrin protein by using In-Silico analysis.
Material and Methods
Patients diagnosed with Steroid resistant nephrotic syndrome were recruited from the Department of pediatric nephrology, Children Hospital, Lahore and the Department of pediatric nephrology, SIUT, Karachi and were subjected to mutation screening of all 29 coding exons of NPHS1 gene as described previously by Abid et al. (2012) [21].
In silico analysis
The observed missense mutations were further analyzed for their potential effects on the protein by using three online pathogenicity predictor tools, SIFT (Sorting Intolerant From Tolerant) (http://sift.jcvi.org/)[22], PROVEAN (Protein Variation Effect Analyzer) (http://provean.jcvi.org/index.php) [23] and Mutation Assessor (http://mutationassessor.org/)[24]. The evolutionary conservation of amino acid where mutations were found, multiple sequence alignment (MSA) across different species was constructed using the CLUSTAL OMEGA tool(http:// www.ebi.ac.uk/Tools/msa/clustalo/) [25,26] and phylogenetic tree was constructed with MEGA 5 program [27]. In an attempt to investigate the potential impact of the NPHS1 mutations on protein structure, in silico analysis was performed using the online protein modeling I-TASSER server (http://zhanglab.ccmb. med.umich.edu/I-TASSER)and were further analyzed by PyMOL Molecular Graphics System, Version 1.7.4.4 2010 Schrodinger, LLC (http://www.pymol.org) for comparison between wild type and mutant protein. Further, translation analysis was done for frame shift and nonsense mutations identified in this study by using Expasy translate tool.
Results and Discussion
Sequence analysis
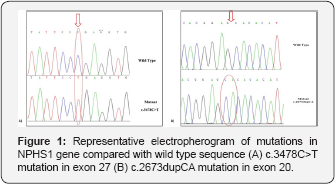
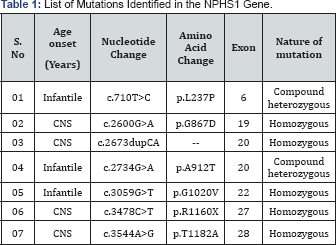
DNA of each individual from recruited patients were amplified and subjected to sequencing for mutational analysis and 7 mutations were identified. Mutations were homozygous or compound heterozygous in nature that includes 5 missense mutations (c.710T>C, c.2600G>A, c.2734G>A, c.3059G>T, c.3544A>G) a nonsense mutation (c.3478C>T) and a frame-shift mutation (c.2673dupCA). Figure 1 shows the representative electropherogram of mutations. Table 1 summarizes the identified mutations with specific nucleotide and amino acid change along with the respective exon. Analysis of missense mutations (c.710T>C, c.2600G>A, c.2734G>A, c.3059G>T, c.3544A>G).
Pathogenicity prediction
In silico analysis was done to predict the potential deleterious effects on protein conformation on the basis of its evolutionary conservation and location in three-dimensional structure of NPHS1 protein. Pathogenicity prediction tools classified three mutations (p.L237P, p.G867D, p.G1020V) as possibly pathogenic/damaging and two (p.A912T, p.T1182A) as a benign/tolerated mutation (Table 2). Homozygous mutation p.G867D was found in a steroid resistant CNS case as a result of consanguineous marriage and parents were segregating the mutant allele in heterozygous state. Although SRNS is known to be a monogenic disease but with the identification of heterozygous (single variants), homozygous, and compound heterozygous mutations, genetic mechanisms in podocytes are becoming increasingly complex. Di-genic inheritance pattern in SRNS patients was previously reported by Abid et al. 2012 and Koziell et al. 2002 [19,21]. According to them di-genic inheritance pattern results in a "tri-allelic" condition, in which patient was found homozygous for a mutation in one gene and heterozygous for a mutation in the other gene. As p.T1182A mutation was found homozygous in nature and predicted as benign so, heterozygous mutation in any other gene may be there for the pathogenesis of disease thus further sequence analysis of other disease causing genes are suggested to confirm the di-genic inheritance pattern in such case. Interestingly in an infantile SRNS patient, the two different heterozygous mutations (p.A912T and p.L237P) that predicted as benign and pathogenic respectively, was found as a compound heterozygous mutation. In this case, parents were carrier for each mutation. This case was possibly showing the evidence for modifier and synergistic effects of genes in SRNS.

aSIFT classify substitutions as damaging (SIFT score: < 0.05), or tolerant (SIFT score: >0.05).
bPROVEAN v1.1: classify substitutions as «deleterious» (PROVEAN score is equal to or below a predefined threshold -2.5), and if the PROVEAN score is above the threshold, the variant is predicted to have a «neutral» effect.
cMutation Assessor gives substitutions as high functional (FI score: 3.50-5.50), Medium functional (FI score: 2.00-3.50), low nonfunctional (FI score: 1.00-2.00) or neutral non-functional (FI score: <1.00)
MSA and phylogenetic tree
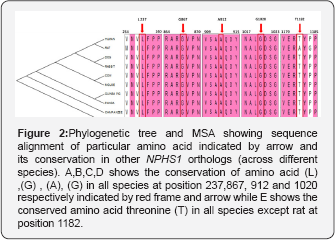
These predictions were also supported by multiple sequence alignment of NPHS1 orthologs. MSA and phylogenetic tree was used as another line of evidence that showed all amino acids, are highly conserved among species from human to zebrafish, except amino acid T1182, which varies in rat where threonine is replaced by alanine (Figure 2). Molecular phylogenetic studies described the fact that highly conserved DNA sequences have functional value which dictates the conservation of amino acid at their specific position and have a very important and crucial role in the integrity of protein structure and conformation of different domains. Through MSA analysis, our study also supports the fact which demonstrated that all these mutations occurred in the codons encoding the amino acids that belongs to a position which has sequence homology among all orthologs. Our results highlight that any mutation at these conserved sites leads to the major disruption in nephrin protein that may cause the disturbance of filtration barrier causing steroid resistant nephrotic syndrome.
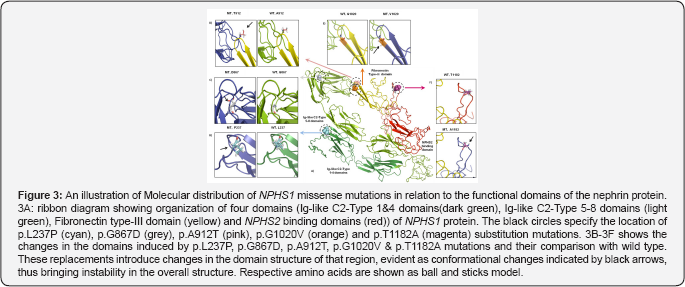
Protein modeling
Structural modeling of missense mutations was done by using the online protein modeling I-TASSER (Figure 3). Protein modeling showed that p.L237P mutation is localized in the Ig -like C2- type 3 domain and represents a situation where leucine, a bigger and nonpolar residue usually buried in folded proteins is replaced by a much smaller and rigid residue, proline. The Ig -like C2- type 3 domain of nephrin protein is an extracellular domain that interacts with NEPH1, a surrounding structural protein. The nephrin-NEPH1 interaction is important in maintaining the architecture and permeability of SD and the substitution in this domain introduce conformational changes which are likely to disrupt nephrin-NEPH1 interaction.
The G867D & A912T mutations are localized in the Ig -like C2- type 8 domain while the G1020V mutation is located at the apex of p-sheet in the fibronectin type III domain. Molecular modeling predicted that these mutations introduce instability in the protein structure through increased steric encumbrance as these domains are involved in interaction with the surrounding molecules in SD. The fifth NPHS1 mutation, p.T1182A affects an amino acid of the unstructured loop in NPHS2 binding domain. The replacement of polar threonine with a smaller non polar alanine is likely to cause rigidity in the structure of the domain. Substitution of alanine in the NPHS2 binding domain at this position not only breaks H-bonds but also cause increased steric hindrance which in turn is predicted to affect the interaction of nephrin protein with podocin.
Analysis of non-sense mutation (c.3478C>T)
Analysis of c.3478C>T, a nonsense mutation revealed that homozygous C to T substitution at nucleotide position c.3478 in exon 27 replaced the codon CGA for arginine (R) with a premature stop codon (TGA) at position (1160). This p.R1160X mutation leads to the synthesis of a truncated protein of 1160 amino acid residues instead of 1241 amino acids. This nonsense mutation result, in the loss of half portion of NPHS2 binding domain resulting in a disruption of nephrin- podocin interaction essential for the functional and structural integrity of glomerular podocytes (28) as represented in Figure 4.

In silico analysis of frame shift mutation (c.2673dupCA)
Translation analysis for homozygous c.2673dupCA mutation was done by using Expasy translate tool (Figure 5). Analysis demonstrated that CA duplication at position 2673 in exon 20 results in shifting of the reading frame from position 893 with the introduction of premature stop codon at position 904 leads to protein truncation. Comparison of crystal diagram with wild type protein model suggested the loss of both fibronectin type III and NPHS2 binding domains in the mutated protein which include some extracellular region, transmembrane and cytosolic region. These domains are responsible for the interaction with surrounding molecules to maintain the integrity of Slit diaphgram. Possibly loss of two these interacting domains may result in over all disruption of filtration barrier leading to proteinuria and steriod resistant nephrotic syndrome.

Conclusion
Through sequence analyses, seven disease associated mutations were found in NPHS1 gene. In-silico analyses showed that 1 mutation is located in the Ig -like C2- type 3 domain, 3 mutations in Ig -like C2- type 8 domain, 1 in fibronectin type III and 2 in NPHS2 binding domains. All of these domains play a critical role during the interaction of nephrin protein with other structural and functional proteins of slit diaphragm. Mutations in these important domains may result in overall disruption of SD. Further, in silico and in-vitro studies needs to be performed to explore the possible effects of these mutations in the structural development and functioning of slit diaphragm.
Acknowledgment
This work was supported by Higher Education Commission of Pakistan grant # 1987 to SK. The authors thank the patients and family members for taking part in this study. We also thank Prof. S Q Mehdi for his guidance and valuable help.
References
- Hinkes BG, Mucha B, Vlangos CN, Gbadegesin R, Liu J, et al. (2007) Nephrotic syndrome in the first year of life: two thirds of cases are caused by mutations in 4 genes (NPHS1, NPHS2, WT1, and LAMB2). Pediatrics 119(4): e907-919.
- Certikova-Chabova V, Tesar V (2013) Recent insights into the pathogenesis of nephrotic syndrome. Minerva medica 104(3): 333-347.
- Avni EF, Vandenhoute K, Devriendt A, Ismaili K, Hackx M, et al. (2011) Update on congenital nephrotic syndromes and the contribution of US. Pediatric radiology 41(1): 76-81.
- Weber S, Gribouval O, Esquivel EL, Moriniere V, Tete MJ, et al. (2004) NPHS2 mutation analysis shows genetic heterogeneity of steroid- resistant nephrotic syndrome and low post-transplant recurrence. Kidney int 66(2): 571-579.
- Trautmann A, Bodria M, Ozaltin F, Gheisari A, Melk A, et al. (2015) Spectrum of steroid-resistant and congenital nephrotic syndrome in children: the PodoNet registry cohort. Clin J Am Soc Nephrol 10(4): 592-600.
- Rennke HG, Cotran RS, Venkatachalam MA (1975) Role of molecular charge in glomerular permeability. Tracer studies with cationized ferritins. J C Biol 67(3): 638-646.
- Karnovsky MJ, Ainsworth SK (1972) The structural basis of glomerular filtration. Advances in nephrology from the Necker Hospital 2: 35-60.
- Rodewald R, Karnovsky MJ (1974) Porous substructure of the glomerular slit diaphragm in the rat and mouse. J C Biol 60(2): 423-33.
- Zenker M, Machuca E, Antignac C (2009) Genetics of nephrotic syndrome: new insights into molecules acting at the glomerular filtration barrier. J Mol Med (Berl) 87(9): 849-857.
- Jalanko H (2009) Congenital nephrotic syndrome. Pediatr Nephrol 24(11): 2121-2128.
- Moudgil A, Nast CC, Bagga A, Wei L, Nurmamet A, et al. (2001) Association of parvovirus B19 infection with idiopathic collapsing glomerulopathy. Kidney Int 59(6): 2126-2133.
- Tomlinson L, Boriskin Y, McPhee I, Holwill S, Rice P (2003) Acute cytomegalovirus infection complicated by collapsing glomerulopathy. Nephrology, dialysis, transplantation. Official publication of the European Dialysis and Transplant Association - European Renal Association 18(1): 187-189.
- Bruggeman LA, Ross MD, Tanji N, Cara A, Dikman S, et al. (2000) Renal epithelium is a previously unrecognized site of HIV-1 infection. J Am Soc Nephrol 11(11): 2079-2087.
- Gee HY, Zhang F, Ashraf S, Kohl S, Sadowski CE, et al. (2015) KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J Clin Invest 125(6): 2375-2384.
- Ruotsalainen V, Ljungberg P, Wartiovaara J, Lenkkeri U, Kestila M, et al. (1999) Nephrin is specifically located at the slit diaphragm of glomerular podocytes. Proc Natl Acad Sci U S A 96(14): 7962-7967.
- Li H, Lemay S, Aoudjit L, Kawachi H, Takano T (2004) SRC-family kinase Fyn phosphorylates the cytoplasmic domain of nephrin and modulates its interaction with podocin. J Am Soc Nephrol 15(12): 3006-3015.
- Pavenstadt H, Kriz W, Kretzler M (2003) Cell biology of the glomerular podocyte. Physiol Rev 83(1):253-307.
- Lenkkeri U, Mannikko M, McCready P, Lamerdin J, Gribouval O, et al. (1999) Structure of the gene for congenital nephrotic syndrome of the finnish type (NPHS1) and characterization of mutations. Am J Hum Genet 64(1): 51-61.
- Koziell A, Grech V, Hussain S, Lee G, Lenkkeri U, et al. (2002) Genotype/ phenotype correlations of NPHS1 and NPHS2 mutations in nephrotic syndrome advocate a functional inter-relationship in glomerular filtration. Hum Mol Genet 11(4): 379-388.
- Bolk S, Puffenberger EG, Hudson J, Morton DH, Chakravarti A (1999) Elevated frequency and allelic heterogeneity of congenital nephrotic syndrome, Finnish type, in the old order Mennonites. Am J Hum Genet 65(6): 1785-1790.
- Abid A, Khaliq S, Shahid S, Lanewala A, Mubarak M, et al. (2012) A spectrum of novel NPHS1 and NPHS2 gene mutations in pediatric nephrotic syndrome patients from Pakistan. Gene 502(2): 133-137.
- Ng PC, Henikoff S (2001) Predicting deleterious amino acid substitutions. Genome Res 11(5): 863-874.
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP (2012) Predicting the functional effect of amino acid substitutions and indels. PloS one 7(10): e46688.
- Reva B, Antipin Y, Sander C (2007) Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol 8(11): R232.
- Ashkenazy H, Kliger Y (2010) Reducing phylogenetic bias in correlated mutation analysis. Protein Eng Des Sel 23(5): 321-326.
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, et al. (2005) ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res 33(Web Server issue): W299-302.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10): 2731-2739.
- Huber TB, Kottgen M, Schilling B, Walz G, Benzing T (2001) Interaction with podocin facilitates nephrin signaling. J of Biol Chem 276(45): 41543-41546.
- Foster RR, Saleem MA, Mathieson PW, Bates DO, Harper SJ (2004) Vascular endothelial growth factor and nephrin interact and reduce apoptosis in human podocytes. Am J Physiol Renal Physiol 288(1):F48-57.






























