Role of MiRNAs in Oral Cancer
Tingting Jia1, Li Wang2, Rui Zhao1, Liang Zhu1, Lejun Xing1 and Bin Guo1*
1Department of stomatology, Medical College of People’s Liberation Army & Chinese People’s Liberation Army General Hospital, China
2Department of Stomatology, Hangzhou Dental Hospital, China
Submission: June 28, 2018; Published: July 12, 2018
*Corresponding author: Bin Guo, Department of stomatology, Chinese PLA General Hospital, 28 Fuxing Road, Beijing 100853, China
How to cite this article: Tingting Jia, Li Wang, Rui Zhao, Liang Z, Lejun X et al. Role of MiRNAs in Oral Cancer. J Head Neck Spine Surg. 2018; 3(2): 555606. DOI: 10.19080/JHNSS.2018.03.555606
Abstract
Oral cancer, one of the most common cancers worldwide, exhibits high mortality and morbidity rates. The incidence rate of oral cancer is high in developing countries, especially in Asian countries. MiRNAs are non-coding RNAs with significant regulatory functions, such as mRNA degradation or translation inhibition. Understanding of the tumorigenesis of oral cancer has significantly progressed at multiple levels. MiRNAs play an important role in oral tumors and have been associated with specific oral cancer phenotypes, such as tumorigenesis, progression, recurrence, or postoperative survival. MiRNAs exist stably in common body fluids and are thus ideal biomarkers for oral cancer. The miRNA profiles hallmark a potential diagnostic value for detection of oral cancer and potentially malignant disorders. In this review, we will summarize our current knowledge regarding the most active miRNAs involved in oral cancer, focusing our discussion on their role in tumor diagnosis, development, and progression.
Keywords:MiRNAs; Oral cancer; Tumorigenesis; Biomarker; Diagnosis; Prognosis
Introduction
Oral cancer, also known as “oral cavity cancer,” is the sixth most common cancer worldwide and has high mortality and morbidity rates. Oral squamous cell carcinoma is the most common type, accounting for approximately 90% of oral cancers [1]. The 5-year survival rate of oral cancer is approximately 50%, and more than 11,000 deaths are recorded annually [2]. Approximately 500,000 individuals are diagnosed annually worldwide, and the incidence rate continuously increases [3]. Oral cancer is prevalent in developing countries, especially in South Asia [2]. Oral cancer is significantly caused by environmental carcinogens, particularly smoking, alcohol, and betel quid chewing [4]. Patients with oral cancer experience poor quality of life because of the side effects of clinical treatments and cosmetic problems. Oral cancer is a multi-step disease, but its precise mechanism remains unknown [5]. Therefore, the pathogenesis of oral cancer and its earlier biomarkers must be further explored.
MiRNAs are non-coding RNAs with approximately 17 to 25 nucleotides in length [6]. The biogenesis of miRNAs begins with transcription of pri-miRNAs by RNA polymerase II. Pri-miRNAs are then cleaved by Drosha in the nucleus, producing pre-miRNAs [6,7]. Pre-miRNAs are subsequently exported into the cytoplasm and processed by Dicer to form mature ∼21bp miRNAs, which are incorporated into RNA-induced silencing complex [6,8]. Mature miRNAs bind to complementary sequences of conserved 3′untranslated regions (UTRs), repressing translation and promoting degradation of target genes (Figure 1) [6,9-11].

In recent decades, miRNAs have been regarded as important factors for tumorigenesis [5,12]. Evidence suggests that miRNAs function as either tumor suppressors or oncogenes [13,14].MiRNAs are differentially expressed in distinct tumors and involved in important tumor-related cellular programs, such as cell proliferation, cell survival, cell differentiation, antiangiogenesis, invasion, or metastasis [15].
Functional MiRNAs in Oral Cancer
Many studies reported that miRNAs are associated with specific oral cancer phenotypes, such as tumorigenesis, progression, recurrence, or postoperative survival [5,16]. In this review, we summarize commonly known miRNAs related to oral cancer and discuss their biological functions or cellular mechanisms [5] (Table 1). In addition, the stable existence of miRNAs in common body fluids makes them ideal biomarkers for diagnosis or prognosis of oral cancer.

Let-7b
Let-7b miRNA, one of the first discovered miRNAs, is a key regulator for cell proliferation and differentiation [17,18]. Let- 7b miRNA is found in many cancers, including colon, pancreatic endocrine, acinar tumors, and pancreatic adenocarcinoma [19]. Andrew et al. [20] found that let-7b, but not let-7a, was significantly reduced in oral cancer cell lines compared with control cells. Over-expression of let-7b significantly reduced the expression of its target gene Dicer, which is an RNase III endonuclease required for miRNA maturation and aberrantly expressed in different types of cancer [20]. Another study found that let-7b also targets insulin-like growth factor 1 receptor (IGF1R), which is activated by IGF1 or IGF2 through autocrine and paracrine signaling. Let-7b inhibits cell proliferation and colony formation and triggers S/G2 cell cycle arrest by targeting IGF1R and IRS-2 through the Akt pathway [1]. Hence, let-7b down-regulation contributes to oral cancer progression. Over-expressing let-7b could be a potential therapy target for inhibition of cancer cell proliferation.
miR-7/miR-21
MiR-7 and miR-21 are two keratinization-associated miRNAs that are up-regulated in keratinized tumors compared with normal or non-keratinized tumors [21]. Jung et al. [22] found that miR-7 and miR-21 are up-regulated; in silico analysis results indicated that RECK is the optimal candidate target gene for miR- 7 and miR-21. RECK plays crucial roles for tumor progression by degrading the extracellular matrix barriers, encompassing the tumor and permitting invasion into surrounding connective tissues. In oral cancer cells, miR-7 and miR-21 are inversely correlated with the expression of RECK [22]. Patricia et al. [23] also found that miR-21 is up-regulated in oral cancer; this study revealed that miR-21 directly binds to the 3′UTR of PDCD4, which is associated with disease progression and metastasis [23]. Further study showed that the expression of miR-21 is the major factor related to the poor prognosis of patients with oral cancer [24]. Overall, miR-7/miR-21-induced deregulation of RECK may contribute to the aggressiveness of tumors.
miR-99a
MiR-99a is characterized as tumor suppressor in several human cancers, including childhood adrenocortical tumors, prostate cancer, liver cancer, head and neck cancer, and oral cancer [25-27]. MiR-99a is one of the most down-regulated miRNAs in oral cancer cell lines compared with normal oral keratinocytes. Transwell and tail vein injection assays indicated that miR-99a functions in tumor migration/invasion and lung colonization. Moreover, target prediction and real-time PCR assay validated that IGF1R is the target gene of miR-99a, which is a transmembrane tyrosine kinase receptor [28]. The expression of miR-99a is negatively correlated with the expression of IGF1R in oral cancer cells. Activation of IGF1R leads to activation of the Ras, Raf, and mitogen-activated protein kinase pathway and thus enhances the proliferation and stimulation of the phosphatidylinositol 3-kinase pathway, resulting in apoptosis inhibition [29]. Collectively, miR-99a mutually regulates its own target, IGF1R, suggesting the possibility of miR-99a for targeting IGF1R in cancer therapy.
miR-100
MiR-100 is down-regulated in many cancers, including ovarian cancer and hepatocellular carcinomas [30,31]. Genetic aberrations are common in oral cancer, and chromosome 11q is among the most common alterations [32,33]. Brian et al. [34] examined the expression of miRNAs mapped to 11q in oral cancer cell lines compared with normal human oral keratinocytes (NHOK); results indicated that mir-100 is down-regulated in oral cancer cells. Cell proliferation is significantly reduced when exogenous miR-100 is over-expressed. Microarray analysis data revealed that over-expression of exogenous miR-100 downregulated a number of target genes including ID1, EGR2, MMP13, and FGFR3; these genes are involved in cell metastasis, myelin development, cell adhesion, and cell growth, respectively. In conclusion, miR-100 inhibits cell proliferation by targeting several key genes involved in cancer and plays an important role in the development and progression of oral cancer [34].
miR-125
MiR-125b is also located in chromosome 11q and involved in important cellular processes, especially in neuronal development and differentiation [33]. The alteration of miR-125b occurs in many cancers, including ovarian cancer, breast cancer, and prostate cancer [35,36]. Consistent with miR-100, miR-125b is also down-regulated in oral cancer cell lines. Over-expression of exogenous miR-125b significantly reduces cell proliferation. In oral cancer, miR-125b regulates the expression of key factors involved in tumorigenesis; these factors include KLF13, CXCL11, and FOXA1. All these target genes have important chemokines and transcription factors, which have crucial biological functions in cell growth, cell proliferation and differentiation, inflammation, angiogenesis, and metastasis [31,34]. Shiiba et al. [16] also found that miR-125b is down-regulated in oral cancer cell lines, resulting in increased cell proliferation rate and decreased radio-sensitivity to X-ray irradiation; this study reported a new target of miR-125b, namely, ICAM2, which is involved in cell proliferation [16]. All these lines of evidence suggest that miR-125b contributes to the development and progression of oral cancer by regulating important target genes. Thus, understanding of miR-125b increases our knowledge of the molecular mechanisms underlying oral cancer. Furthermore, miR-125b may be a potential therapeutic target for oral cancer.
miR-145
MiR-145 acts a tumor suppressor in numerous human cancers, including prostate cancer, bladder cancer, and colon cancer [37-39]. A study found that miR-145 is down-regulated in oral cancer tissues compared with normal mucosa tissues. Exogenous over-expression of miR-145 induced G1 phase arrest and cell apoptosis, which then suppressed cell proliferation and colony formation. The two direct targets of miR-145 are c-Myc and Cdk6, which are important oncogenes. In oral cancer cells, miR-145 also targets c-Myc and Cdk6, leading to inhibition of oral cancer cell growth [40]. Thus, miR-145 rescue may be a rationale for diagnostic and therapeutic applications in oral cancer.
miR-146a
MiR-146a is suppressed in many malignancies [41] but has been recently identified to be up-regulated in oral cancer [42]. Hung et al. [43] found that miR-146a exhibits higher expression in oral cancer than matched adjacent mucosal cells. Inhibition of miR-146a significantly blocks the growth of xenograft tumors. miR-146a directly targets IRAK1, TRAF6, and NUMB, thereby enhancing cell proliferation, invasion, and metastasis. The plasma miR-146a levels of patients with oral cancer are significantly higher than those of control subjects; hence, miR-146a could be a potential biomarker for diagnosis of oral cancer [43].
miR-155
MiR-155 is overexpressed in many human cancers, including lymphoma and solid tumors of diverse origin (e.g., breast, lung, stomach, prostrate, colon, thyroid, and pancreatic) [14,44,45]. MiR-155 is also up-regulated in oral cancer [46]. In oral cancer, miR-155 targets CDC73, which negatively regulates β-catenin, cyclin D1, and c-MYC. Ectopic expression of miR-155 dramatically reduced CDC73 levels, increased cell viability, and decreased apoptosis in nude mice. Conversely, inhibiting miR- 155 resulted in increased CDC73 levels, decreased cell viability, and increased apoptosis. Hence, the tumor suppressor CDC73 is a target of oncogenic miR-155 [47]. Furthermore, the reversal of pro-oncogenic properties of miR-155 due to its inhibitor is a promising method for cancer therapeutics [47].
miR-196
MiR-196 is an active molecule and is altered in several cancers. Studies stated that miR-196 is down-regulated in melanoma and acute lymphoblastic leukemia [48]. However, other scholars reported that miR-196 is up-regulated in several malignant diseases, including esophageal cancer, pancreatic cancer, colorectal cancer, and glioblastoma [49]. Lu et al. [50] examined the expression of miR-196 in oral cancer cell lines and normal keratinocyte cell lines; results indicated that miR-196 is highly up-regulated in oral cancer cells. In vitro experiments showed that miR-196 is responsible for cell growth, migration, invasion, and radio/chemosensitivity. Target prediction and subsequent validation experiments identified that miR-196 targets NME4, a member of the NM23 family. NME4 then regulates the downstream JNK-TIMP1-MMP signaling pathway, which plays an important role in cell mobility [50]. In conclusion, miR- 196 promotes invasive and migratory phenotypes in oral cancer by targeting NME4, leading to the regulation of the downstream JNK-TIMP1-MMP signaling pathway [51].
miR-205
Various studies examined the biological functions of microRNA-205 (miR-205) as a tumor suppressor in carcinogenesis [52]. Kim et al. [53] found that miR-205 is significantly down-regulated in human oral cancer compared with NHOKs. Reduced miR-205 expression in oral cancer cells might be important for oral cancer progression. Over-expressing exogenous miR-205 reduced cell viability in oral cancer cells and induced cell apoptosis by activating caspase-3/-7. In addition to caspase-3/-7, over-expressed miR-205 significantly promotes the expression of IL-24, a well-known cytokine that functions in cell apoptosis. IL-24 promoter scanning analysis revealed a potential binding sequence located at position-151 in the IL-24 promoter. miR-205 interacts directly with the promoter target sequence of IL-24 [53]. Some studies stated that miRNA can upregulate the expression of their own target genes through an interaction with the regulatory region [54,55]. These findings indicate that miR-205 functions as a tumor suppressor in upregulating IL-24 by targeting specific sites in its promoter. Thus, miR-205 exhibits a significant therapeutic potential as molecular medicine for treatment of oral cancer.
miR-483-3p
MiR-483-3p is involved in the final stage of skin wound healing and arrest of keratinocyte proliferation [56]. MiR-483-3p is down-regulated in oral cancer cells. Over-expressing miR-483- 3p significantly hinders tumor growth. Target prediction analysis and validation experiments revealed that miR-483-3p targets API5, RAN, and BIRC5, which play a main role in cell apoptosis. MiR-483-3p exhibits its pro-apoptotic activity by binding to these target genes, leading to inhibition of cell proliferation and increase in cell apoptosis [57]. Hence, miR-483-3p may be used as an adjuvant in many cancers characterized by downregulation of miR-483-3p.
miR-518c-5p
MiR-518c-5p belongs to the miR-515 family and is located in chromosome 19. MiR-518c-5p is originally identified in 26 different organ systems of humans and rodents and is enriched in neuronal as well as normal and malignant hematopoietic cells and tissues [58]. However, the function of miR-518c-5p in cancers remains unclear. Makoto et al. [59] found that SDF-1/ CXCR4 system is involved in the establishment of metastasis in oral cancer. Subsequent examination showed that miR-518c- 5p is induced and exhibits distant metastatic potential. When inhibiting the function of miR-518c-5p by using LNA-modified inhibitors, the cell growth and migration of oral cells are reduced significantly. Consistently, over-expression of miR-518c-5p enhances the migration of oral cancers. An in vivo experiment indicated that exogenous miR-518c-5p significantly increased the tumor volume, lymph node metastasis, and lung metastasis in mice. The miR-518c-5p inhibitor may serve as an ideal target for development of therapies against metastasis [59]. However, the target genes of miR-518c-5p remain unknown. Future studies must critically examine the target genes of miR-518c-5p in oral cancer.
Biomarker MiRNAs for Oral Cancer
Early detection of potentially malignant oral cancer is important for improving the probability of complete recovery because the stage of malignancy at the time of diagnosis influences morbidity and mortality [60]. However, more than 60% of patients present with stage III and IV oral cancer. As such, scholars must explore the most potential biomarkers for early detection of oral cancer (Table 2) [60].
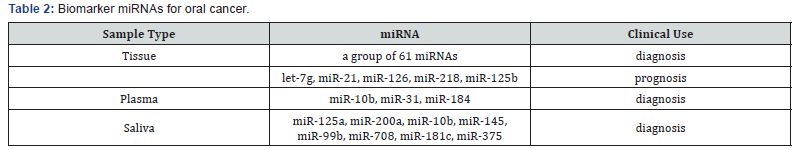
Lajer et al. [61] compared 51 patients with oral cancer with 40 control patients by using microarray analysis and found 114 differentially expressed miRNAs. A molecular classifier including 61 miRNAs was generated for diagnosis of oral cancer; this classifier exhibits 93% accuracy, 100% sensitivity, and 86% specificity [61].
Peng et al. [62] examined the profile of miRNA from 58 oral cancer samples and paired normal tissues through microarray assay. A total of 232 of the 760 miRNAs assessed were differentially expressed between paired tumor and normal tissue samples. Among the 232 miRNAs, the reduced expression of miR-218, miR-125b, and let-7g is associated with high risk of poor outcomes [62]. MiR-21 is a common biomarker for many cancers. A previous study on 86 patients with oral cancer revealed that miR-21 was primarily expressed in the tumor stroma but was not expressed in the adjacent normal epithelia. Multivariate Cox-regression analysis of disease-free survival revealed that miR-21 expression level was significantly associated with poor prognosis [63]. Sasahira et al. [64] found that decreased miR-126 expression was strongly correlated with disease-free survival in 118 cases with oral cancer. Another multivariate analysis of patients with oral cancer indicated that a poor prognosis was associated with low miR-126 expression compared with high miR-126 expression (P=0.0013) [64].
Circulating Biomarker MiRNAs for Oral Cancer
Some biological molecules are present in body fluids, such as plasma and saliva, and are ideal noninvasive biomarkers for oral cancer [65]. Previous studies used protein, mRNA, and DNA extracted from body fluids to detect oral cancer. Analysis using transcriptomic and proteomic technology discovered and validated mRNA and protein salivary biomarkers, respectively, to be highly discriminatory for oral cancer detection. Plasmic and salivary miRNAs are protected from ribonucleases present in the plasma/saliva by macromolecules called exosomes, which are known to package and transport miRNAs [65-67]. The presence of miRNAs in human body fluids, especially in saliva, is an emerging field for monitoring oral diseases.
Liu et al. [68] identified that miR-31 in plasma was significantly elevated in patients with oral cancer (n = 43) compared with control individuals (n = 21). Plasma miR-31 yielded a receiver operating characteristic curve area of 0.82 and an accuracy of 0.72 defined by leave-one-out cross-validation [68].
Wong et al. [69] profiled the expression of 156 miRNAs on four oral cancer and paired normal tissues, revealing 37 differentially expressed miRNAs (three-fold). Among the 37 miRNAs, miR-184 was further examined in the plasma from 20 patients with oral cancer and normal individuals. Plasma miR- 184 level was significantly higher in patients with oral cancer and s, and significantly decreased after surgical removal of the primary tumors [69]. Lu et al. [70] found that miR-10b was considerably elevated in the plasma of patients with oral cancer [P < 0.0001, area under curve (AUC) = 0.932] and pre-cancer lesions (P < 0.0001, AUC = 0.967); hence, miR-10b could be a potential early diagnostic biomarker for oral cancer [70].
Park et al. [66] examined and compared the expression of approximately 50 miRNAs in whole saliva and saliva supernatant between patients with oral cancer and healthy controls. Two saliva miRNAs, namely, miR-125a and miR-200a, were present in significantly lower levels (P<0.05) in the patients than that in the controls [66]. Yang et al. [71] first examined the profile of saliva miRNAs in 10 patients who developed carcinoma and 35 patients who did not develop oral cancer; results indicated that miR-10b, miR-145, miR-99b, miR-708 and miR-181c were significantly differentially expressed [71]. Wiklund et al. [72] used TaqMan RT-PCR arrays to profile miRNA expression in saliva from 15 patients with oral cancer and 7 healthy controls; the results showed that miR-375 was down-regulated in the patients and thus could be used to diagnose oral cancer [72].
Conclusion
Oral cancer is the sixth most common cancer worldwide and has high mortality and morbidity rates. Understanding the tumorigenesis and development of oral cancer has significantly progressed. The expression of many miRNAs is altered in oral cancer, thereby regulating cell growth, cell proliferation and apoptosis, invasion, and metastasis. Accumulating evidence highlights that circulating miRNAs are significantly associated with oral cancer and thus could be used as potential biomarkers for diagnosis and prognosis of oral cancer [73-75]. However, the highly stability makes miRNA ideal biomarkers for non-invasion detection. Further work is required to elucidate the complete regulatory mechanisms of miRNAs. The clinical use of miRNA as biomarkers needs more tests and more patients to validate the most potential markers.
References
- Gao L, Wang X, Wang X, Zhang L, Qiang C, et al. (2014) IGF-1R, a target of let-7b, mediates crosstalk between IRS-2/Akt and MAPK pathways to promote proliferation of oral squamous cell carcinoma. Oncotarget 5(9): 2562-2574.
- Adami GR, Tangney CC, Tang JL, Zhou Y, Ghaffari S, et al. (2018) Effects of green tea on miRNA and microbiome of oral epithelium. Scientific Reports 8(5873).
- Ahmadi-Motamayel F, Bayat Z, Hajilooi M, Shahryar-Hesami S, Mahdavinezhad A, et al. (2017) Evaluation of the miRNA-146a and miRNA-155 Expression Levels in Patients with Oral Lichen Planus. Iran J Immunol 14(4): 316-324.
- Arunkumar G, Ak DMR, Manikandan M, Rao HPS, Subbiah S, et al. (2018) Dysregulation of miR-200 family microRNAs and epithelialmesenchymal transition markers in oral squamous cell carcinoma. Oncol Lett 15(1): 649-657.
- Gasche JA, Goel A (2012) Epigenetic mechanisms in oral carcinogenesis. Future Oncol 8(11): 1407-1425.
- Binmadi NO, Basile JR, Perez P, Gallo A, Tandon M, et al. (2017) miRNA Expression Profile of Mucoepidermoid Carcinoma. Oral Dis 24(4): 537- 543.
- Chang YA, Weng SL, Yang SF, Chou CH, Huang WC, et al. (2018) A Three- MicroRNA Signature as a Potential Biomarker for the Early Detection of Oral Cancer. Int J Mol Sci 19(3): 758.
- Wostenberg C, Lary JW, Sahu D, Acevedo R, Quarles KA, et al. (2012) The role of human Dicer-dsRBD in processing small regulatory RNAs. PLoS One 7(12): e51829.
- Schneider A, Victoria B, Lopez YN, Suchorska W, Barczak et al. (2018) Tissue and serum microRNA profile of oral squamous cell carcinoma patients. Scientific Reports 8(675).
- Ding Y, Liu P, Zhang S, Tao L, Han J (2018) Screening pathogenic genes in oral squamous cell carcinoma based on the mRNA expression microarray data. Int J Mol Med 41(6): 3597-3603.
- Dumache R (2017) Early Diagnosis of Oral Squamous Cell Carcinoma by Salivary microRNAs. Clin Lab 63(11): 1771-1776.
- Shiiba M, Uzawa K and Tanzawa H (2010) MicroRNAs in head and neck squamous cell carcinoma (HNSCC) and oral squamous cell carcinoma (OSCC). Cancers (Basel) 2(2): 653-669.
- El-Sakka H, Kujan O, Farah CS (2018) Assessing miRNAs profile expression as a risk stratification biomarker in oral potentially malignant disorders: A systematic review. Oral Oncol 77: 57-82.
- Fang Z, Zhao J, Xie W, Sun Q, Wang H, et al. (2017) LncRNA UCA1 promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by sunppressing miR‐184 expression. Cancer Med 6(12): 2897-2908.
- Farah CS, Fox SA, Dalley AJ (2018) Integrated miRNA-mRNA spatial signature for oral squamous cell carcinoma: a prospective profiling study of Narrow Band Imaging guided resection. Sci Rep 8(823).
- Shiiba M, Shinozuka K, Saito K, Fushimi K, Kasamatsu A, et al. (2013) MicroRNA-125b regulates proliferation and radioresistance of oral squamous cell carcinoma. Br J Cancer 108(9): 1817-1821.
- Gai C, Camussi F, Broccoletti R, Gambino A, Cabras M, et al. (2018) Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. BMC Cancer 18(1): 439.
- Ikehata N, Takanashi M, Satomi T, Watanabe M, Hasegawa O, et al. (2017) Toll-like receptor 2 activation implicated in oral squamous cell carcinoma development. Biochem Biophys Res Commun 495(3): 2227- 2234.
- Ikoma M, Gantt S, Casper C, Ogata Y, Zhang Q, et al. (2018) KSHV oral shedding and plasma viremia result in significant changes in the extracellular tumorigenic miRNA expression profile in individuals infected with the malaria parasite. Plos One 13(2): e0192659.
- Jakymiw A, Patel RS, Deming N, Bhattacharyya I, Shah P, et al. (2010) Overexpression of dicer as a result of reduced let‐7 MicroRNA levels contributes to increased cell proliferation of oral cancer cells. Genes Chromosomes Cancer 49(6): 549-559.
- Mehdipour M, Shahidi M, Manifar S, et al. (2018) Diagnostic and prognostic relevance of salivary microRNA-21, -125a, -31 and -200a levels in patients with oral lichen planus - a short report. Cellular Oncology p. 1-6.
- Jung HM, Phillips BL, Patel RS, Cohen DM, Jakymiw A, et al. (2012) Keratinization-associated miR-7 and miR-21 regulate tumor suppressor reversion-inducing cysteine-rich protein with kazal motifs (RECK) in oral cancer. J Biol Chem 287(35): 29261-29272.
- Shah S, Jadhav K, Shah V, Gupta N, Dagrus K (2016) miRNA 21: Diagnostic Prognostic and Therapeutic marker for Oral Cancer. Microrna 5(3): 175-179.
- Reis PP, Tomenson M, Cervigne NK, Machado J, Jurisica I, et al. (2010) Programmed cell death 4 loss increases tumor cell invasion and is regulated by miR-21 in oral squamous cell carcinoma. Mol Cancer 9: 238.
- Doghman M, El Wakil A, Cardinaud B, Thomas E, Wang J, et al. (2010) Regulation of insulin-like growth factor–mammalian target of rapamycin signaling by microRNA in childhood adrenocortical tumors. Cancer Res 70(11): 4666-4675.
- Sun D, Lee YS, Malhotra A, Kim HK, Matecic M, et al. (2011) miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res 71(4): 1313- 1324.
- Li D, Liu X, Lin L, Hou J, Li N, et al. (2011) MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. Journal of Biological Chemistry 286(42): 36677-36685.
- Martelli F, Mencarini J, Rocca A, Malva ND, Bartolozzi D, et al. (2018) Polyomavirus microRNA in saliva reveals persistent infectious status in the oral cavity. Virus Res 249: 1-7.
- Yen YC, Shiah SG, Chu HC, Hsu YM, Hsiao JR, et al. (2014) Reciprocal regulation of MicroRNA-99a and insulin-like growth factor I receptor signaling in oral squamous cell carcinoma cells. Mol Cancer 13: 6.
- Nagai H, Hasegawa S, Uchida F, Terabe T, Ishibashi Kanno N, et al. (2018) MicroRNA-205-5p suppresses the invasiveness of oral squamous cell carcinoma by inhibiting TIMP 2 expression. Int J Oncol 52(3): 841-850.
- Dossantos MF, Dasilva AFM, Carvalho ACPD, Pereire CM, Sehnem D, et al. (2017) miRNAs: Important Targets for Oral Cancer Pain Research. BioMed Research International 2017: 1-8.
- Sannigrahi MK, Sharma R, Singh V, Panda NK, Rattan V, et al. (2018) DNA methylation regulated microRNAs in HPV-16-induced head and neck squamous cell carcinoma (HNSCC). Mol Cell Biochem.
- Santos PRB, Coutinho-Camillo CM, Soares FA, Freitas VS, Vilas-Bôas DS, et al. (2017) MicroRNAs expression pattern related to mast cell activation and angiogenesis in paraffin-embedded salivary gland tumors. Pathol Res Pract 213(12): 1470-1476.
- Sun C, Li J (2018) Expression of MiRNA-137 in oral squamous cell carcinoma and its clinical significance J BUON 23(1): 167-172.
- Shang A, Lu WY, Yang M, Zhou C, Zhang H, et al. (2017) miR-9 induces cell arrest and apoptosis of oral squamous cell carcinoma via CDK 4/6 pathway. Artif Cells Nanomed Biotechnol 26: 1-9.
- Cao M, Zheng L, Liu J, Dobleman T, Hu S, et al. (2018) MicroRNAs as effective surrogate biomarkers for early diagnosis of oral cancer. Clin Oral Investig 22(2): 571-581.
- Tandon D, Dewangan J, Srivastava S, Garg VK, Rath SK (2017) miRNA genetic variants: As potential diagnostic biomarkers for oral cancer. Pathol Res Pract 214(2): 281-289.
- Troiano G, Mastrangelo F, Caponio VCA, Laino L, Cirillo N, et al. (2018) Predictive Prognostic Value of Tissue-Based MicroRNA Expression in Oral Squamous Cell Carcinoma: A Systematic Review and Metaanalysis. J Dent Res 97(7): 759-766.
- Zahra A, Rubab I, Malik S, Khan A, Khan MJ, et al. (2018) Meta-Analysis of miRNAs and Their Involvement as Biomarkers in Oral Cancers. BioMed Research International 2018(8439820): 9.
- Shao Y, Qu Y, Dang S, Yao B, Ji M (2013) MiR-145 inhibits oral squamous cell carcinoma (OSCC) cell growth by targeting c-Myc and Cdk6. Cancer Cell Int 13(1): 51.
- Zeljic K, Jovanovic I, Jovanovic J, Magic Z, Stankovic A, et al. (2018) MicroRNA meta-signature of oral cancer: evidence from a metaanalysis. Ups J Med Sci 123(1): 43-49.
- Slouka Z, Senapati S, Shah S, Lawler R, Shi Z, et al. (2015) Integrated, DC voltage-driven nucleic acid diagnostic platform for real sample analysis: Detection of oral cancer. Talanta 145: 35-42.
- Hung PS, Liu CJ, Chou CS, Kao SY, Yang CC, et al. miR-146a Enhances the Oncogenicity of Oral Carcinoma by Concomitant Targeting of the IRAK1, TRAF6 and NUMB Genes. PloS One 8(11): e79926.
- Zeng J, Yi X, Liu H, Yang Y, Duan Y, et al. Polymorphisms in four microRNAs and risk of oral squamous cell cancer: a meta-analysis. Oncotarget 9(9): 8695-8705.
- Zhang H, Li T, Zheng L, Huang X (2017) Corrigendum to “Biomarker MicroRNAs for Diagnosis of Oral Squamous Cell Carcinoma Identified Based on Gene Expression Data and MicroRNA-mRNA Network Analysis”. Comput Math Methods Med 2017: 1284606.
- Baba O, Hasegawa S, Nagai H, Uchida F, Yamatoji M, et al. (2016) MicroRNA-155-5p is associated with oral squamous cell carcinoma metastasis and poor prognosis. J Oral Pathol Med 45(4): 248-255.
- Rather MI, Nagashri MN, Swamy SS, Gopinath KS, Kumar A (2013) Oncogenic MicroRNA-155 Down-regulates Tumor Suppressor CDC73 and Promotes Oral Squamous Cell Carcinoma Cell Proliferation Implications for Cancer Therapeutics. J Biol Chem 288(1): 608-618.
- Mueller DW, Bosserhoff AK (2011) MicroRNA miR‐196a controls melanoma‐associated genes by regulating HOX‐C8 expression. Int J Cancer 129(5): 1064-1074.
- Haiart S, Watson DI, Leong MP, Astill D, Bright T, et al. (2011) MicroRNA- 196a & microRNA-101 expression in Barrett’s oesophagus in patients with medically and surgically treated gastro-oesophageal reflux. BMC Res Notes 4: 41.
- Keshet Y, Seger R (2010) The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol Biol 661: 3-38.
- Lu YC, Chang JT, Liao CT, Kang CJ, Huang SF, et al. (2104) OncomiR-196 promotes an invasive phenotype in oral cancer through the NME4-JNKTIMP1- MMP signaling pathway. Mol Cancer 13: 218.
- Zheng H, Li S (2017) Reduced miRNA 214 expression in oral mucosa contributes to the pathogenesis of oral lichen planus by targeting CD44. Mol Med Rep 17(1): 1919-1925.
- Kim JS, Yu SK, Lee MH, Park MG, Park E, et al. (2013) MicroRNA-205 directly regulates the tumor suppressor, interleukin-24, in human KB oral cancer cells. Mol Cells 35(1): 17-24
- Bhatnagar N, Li X, Padi SK, Zhang Q, Tang MS, et al. (2010) Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis 1: e105.
- Majid S, Dar AA, Saini S, Yamamura S, Hirata H, et al. (2010) MicroRNA‐205–directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer 116(24): 5637-5649.
- Bertero T, Gastaldi C, Bourget-Ponzio I, Imbert V, Loubat A, et al. (2011) miR-483-3p controls proliferation in wounded epithelial cells. FASEB J 25(9): 3092-3105.
- Bertero T, Bourget-Ponzio I, Puissant A, Loubat A, Mari B, et al. (2013) Tumor suppressor function of miR-483-3p on squamous cell carcinomas due to its pro-apoptotic properties. Cell Cycle 12(14): 2183-2193.
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, et al. (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129(7): 1401-1414
- Kinouchi M, Uchida D, Kuribayashi N, Tamatani T, Nagai H et al. (2014) Involvement of miR-518c-5p to growth and metastasis in oral cancer. PloS One 9(12): e115936.
- Kujan O, Glenny AM, Oliver R, Thakker N, Sloan P (2006) Screening programmes for the early detection and prevention of oral cancer. Cochrane Database Syst Rev 3: CD004150.
- Lajer CB, Nielsen FC, Friis-Hansen L, Norrild B, Borup R, et al. (2011) Different miRNA signatures of oral and pharyngeal squamous cell carcinomas: a prospective translational study. Br J Cancer 104(5): 830- 840.
- Peng SC, Liao CT, Peng CH, Cheng AJ, Chen SJ, et al. MicroRNAs MiR-218, MiR-125b, and Let-7g Predict Prognosis in Patients with Oral Cavity Squamous Cell Carcinoma. PloS One 9(7): e102403.
- Hedbäck N, Jensen DH, Specht L, Fiehn AM, Therkildsen MH, et al. (2014) MiR-21 Expression in the Tumor Stroma of Oral Squamous Cell Carcinoma: An Independent Biomarker of Disease Free Survival. PloS one 9(4): e95193.
- Sasahira T, Kurihara M, Bhawal UK, Ueda N, Shimomoto T, et al. (2012) Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br J Cancer 107(4): 700-706.
- Yoshizawa JM, Wong DT (2013) Salivary microRNAs and oral cancer detection. Methods Mol Biol 936: 313-324.
- Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, et al. (2009) Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res 15(17): 5473-5477.
- Palanisamy V, Sharma S, Deshpande A, Zhou H, Gimzewski J, et al. (2010) Nanostructural and transcriptomic analyses of human saliva derived exosomes. PloS One 5(1): e8577.
- Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW, et al. (2010) Increase of microRNA miR‐31 level in plasma could be a potential marker of oral cancer. Oral Dis 16(4): 360-364.
- Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, et al. (2008) Mature miR- 184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res 14(9): 2588-2592.
- Lu YC, Chen YJ, Wang HM, Tsai CY, Chen WH, et al. (2012) Oncogenic function and early detection potential of miRNA-10b in oral cancer as identified by microRNA profiling. Cancer Prev Res (Phila) 5(4): 665- 674.
- Yang Y, Li YX, Yang X, Jiang L, Zhou ZJ, et al. (2013) Progress risk assessment of oral premalignant lesions with saliva miRNA analysis. BMC cancer 13: 129.
- Wiklund ED, Gao S, Hulf T, Sibbritt T, Nair S, et al. (2011) MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PLoS One 6(11): e27840.
- Ren Y, Zhu H, Chi C, Yang F, Xu X (2015) MiRNA-139 regulates oral cancer Tca8113 cells apoptosis through Akt signaling pathway. Int J Clin Exp Pathol 8(5): 4588-4594.
- Roy R, Singh R, Chattopadhyay E, Ray A, Sarkar ND, et al. (2016) MicroRNA and target gene expression based clustering of oral cancer, precancer and normal tissues. Gene 593(1): 58-63.
- Tian X, Chen Z, Shi S, Wang X, Wang W, et al. (2015) Clinical Diagnostic Implications of Body Fluid MiRNA in Oral Squamous Cell Carcinoma: A Meta-Analysis. Medicine (Baltimore) 94(37): e1324.