VDR Gene Haplotypes Associate with Type 1 Diabetes and Suggest Interaction with HLA, IL2RA and CTLA4 loci in Colombian Families
Natalia Gomez Lopera1, Alejandra Rodriguez1, Martin Toro2, Maria Victoria Lopera2, Alejandra Velez3, Juan Manuel Alfaro1,2 and Nicolas Pineda Trujillo1*
1Grupo Mapeo Genético, Departamento de Pediatría, Facultad de Medicina. Universidad de Antioquia, Colombia
2Seccion de Endocrinología, Departamento de Pediatria, Universidad de Antioquia, Colombia
3Pontificia Universidad Bolivariana, Facultad de Medicina, Colombia
Submission: December 22, 2017; Published: March 14, 2018
*Corresponding author: Nicolas Pineda Trujillo, Grupo Mapeo Genetico, Departamento de Pediatria, Facultad de Medicina, Universidad de Antioquia, Medellín, Carrera 51 D # 62-29, Colombia, Tel: +574 2196065; Email: nicolas.pineda@udea.edu.co
How to cite this article: Natalia G L, Alejandra R, Martin T, Maria V L, Alejandra V et.al. VDR Gene Haplotypes Associate with Type 1 Diabetes and Suggest Interaction with HLA, IL2RA and CTLA4 loci in Colombian Families. Curre Res Diabetes & Obes J. 2018; 6(3): 555688. DOI: 10.19080/CRDOJ.2018.06.555688.
Abstract
Vitamin D and its receptor are possibly related to T-cell-mediated autoimmune diseases and influences susceptibility to Type 1 diabetes (T1D). The aim of this study was to assess the contribution of vitamin D receptor (VDR) polymorphisms to the susceptibility to T1D in a Colombian sample. Associations of VDR gene variants with T1D were investigated in a sample of 200 familial trios from Colombia. VDR polymorphisms FokI (rs2228570), BsmI (rs1544410), ApaI (rs7975232) and TaqI (rs731236), were determined using PCR-RFLP methodology. For interaction tests, we used data obtained in this sample for other T1D candidate loci, such as HLA, PTPN22, INS, IL2RA, SUMO4, CLEC16A, CTLA4 and IFIH1 genes. Allele and genotypic transmission disequilibrium tests were performed; haplotypes and gene-gene interactions were also analyzed. For characterization of the sample, the probands were tested for autoantibodies against tyrosine phosphatase-like 2 (IA-2) and anti-glutamic acid decarboxylase 65 (GAD65). We found that two haplotypes at loci BsmI-ApaI-TaqI have a protective effect in our sample [Haplotypes A-G-T OR=0.219, ci95% (0.12-0.519), p-value=5.326x10-5, and G-T-C OR=0.376, ci95% (0.18-0.786)]. In addition, locus BsmI shows a trend of interaction with SNP rs3087243 (at CTLA4 gene, p-value=8x10-3). Likewise, it was observed a suggestion of interaction of VDR variants with IL2RA, HLA-I and HLA-II. In conclusion, this family-based study presents evidence of two haplotypes and interaction of VDR variants with other loci associated with T1D in Colombian families.
Keywords: VDR gene; Interaction; Haplotype; Colombian families
Introduction
Type 1 diabetes (T1D) is a complex disease characterized by autoimmune and progressive destruction of insulin-secreting pancreatic beta-cells. It is well known that T1D is caused by complex interactions between many genetic and environmental factors [1]. The most firmly established known genetic contribution to T1D susceptibility has been the human leukocyte antigen region (HLA) on the short arm of chromosome 6. However, several other non-HLA regions have been identified as predisposing factors [2]. Among these susceptibility genes, the VDR gene is one of the most widely studied and increasing evidence suggests that vitamin D and its receptor are possibly related to T-cell-mediated autoimmune disease and influence susceptibility to T1D [3].
During the last years, diverse studies have emerged to analyze a possible relationship between T1D and polymorphisms in the Vitamin D receptor (VDR), yielding conflicting results; some of them showed significant association while others failed to reach statistical significance. A more recent meta-analysis found that BsmI and FokI polymorphisms are associated with a significantly increased risk of T1D [4], whereas the ApaI, and TaqI polymorphisms do not appear to have a significant association with overall T1D risk. However, in a later meta-analysis, Tizaou et al. [5] found that individual VDR polymorphisms seemed not to be associated with T1D risk. However, they found that haplotypes contributed significantly to disease susceptibility.
Vitamin D exerts its effects through the vitamin D receptor (VDR). The VDR is found in almost all cells of the immune system, especially the antigen-presenting cells (macrophages and dendritic cells), activated T lymphocytes, as well as in pancreatic islet beta-cells [6]. It is acknowledged that Vitamin D is an effective immunosuppressant via inhibition of lymphocyte activation and cytokine production and prevents or markedly suppresses the development of several autoimmune diseases in animal models [7]. For example, at the level of the pancreatic islets, vitamin D decreased in-vivo and in-vitro proinflammatory chemokine and cytokine expression (e.g., IL6), which are implicated in the pathogenesis of T1D, making beta-cells less chemo attractive and less prone to inflammation [6]. Furthermore, Vitamin D decreases HLA class I expression leading to reduced vulnerability of islet Beta-cells to cytotoxic T lymphocytes [8].
The human VDR gene is located on chromosome 12q13.11, spanning 63.495 bp and includes eight protein-coding exons (exons 2-9) and six untranslated exons (exons 1a-1f), which are alternatively spliced [9]. Four common single nucleotide polymorphisms (SNPs) in the VDR gene have been investigated extensively: FokI C>T (rs2228570), BsmI G>A (rs1544410), ApaI T>G (rs7975232), and TaqI T>C (rs731236) [10] Allele T of the FokI SNP creates an alternative ATG initiation codon in exon 2 leading to a three amino acids longer of the VDR protein. The shorter form of the protein (424 aa) is more active than the long form (427 aa). The BsmI and ApaI SNPs are both located in intron 8, and the Taq I is a silent SNP in exon 9. These three polymorphisms lead to silent variants associated with increased VDR mRNA stability [11].
The above apparent discrepancies among different studies could be a result of the effect of ethnic differences related to the distribution of VDR polymorphisms amongst populations, as well as to interactions with other genetic or environmental factors involved in the pathogenesis of type 1 diabetes [12]. These discrepancies demonstrate that the role of VDR polymorphisms in T1D pathogenesis is unclear. For this reason, the aim of this study was to assess the contribution of vitamin D receptor polymorphisms to the susceptibility to T1D in a Colombian population. Our results may help clarify whether VDR gene variants contribute or not to the susceptibility for being affected by T1D. Also, they could contribute to elucidating whether the role of such variants is allelic or genotypic.
Patients and methods
Subjects
The subjects for the present study are from Antioquia, Colombia. This population corresponds to a genetically special population, described as a mixed population with linkage disequilibrium sources and founder effects [13-15], which makes it ideal for mapping complex traits, such as T1D. The study group consists of 200 trios (two parents and their affected child). The cases are children diagnosed with T1D before 15 years old.
Patients were identified in the Pediatric Endocrinology Program (Universidad de Antioquia and Hospital San Vicente Fundación), IPS universitaria-UdeA, Universidad Pontificia Bolivariana, and Instituto Antioqueño de Diabetes. This study was approved by the ethics committee of the Medicine school of Universidad de Antioquia, and informed consent was obtained from patients and their parents prior blood sampling.
Inclusion criteria were as follows: 1) At least six greatgrandparents of affected children originated in any of the Colombian states of Antioquia, Risaralda, Quindío, Caldas, North of Cauca Valley, North of Tolima, which indicates self-reported “Paisa” ancestry. 2) Both parents willing to participate in the study.
DNA isolation and VDR genotyping
Genomic DNA was isolated from EDTA whole peripheral blood samples using phenol-chloroform protocol [16]. Genotypes for FokI (rs2228570), BsmI (rs1544410), ApaI (rs7975232) and TaqI (rs731236) of VDR gene were obtained by DNA amplifications with standard PCR and specific sets of primers [17], followed by restriction fragment length polymorphism method (RFLP). All amplifications were carried out in a total volume of 15uL, containing 30ng of purified DNA, 1.5μL 10X reaction buffer containing Tris-Hcl, 10mM of each dNTPs, 100nM of each primer and 1.15 U of Taq Polymerase; 1.5mM of MgCl2 for FokI, TaqI and ApaI; and 2.0mM for BsmI. All PCRs were done in a BioRad Diad thermal cycler.
PCR products were analyzed on 2% agarose gels containing 5mg/mL ethidium bromide and visualized under a UV transilluminator. The amplified products were digested using the restriction enzymes BsmI, FokI, ApaI and TaqI, according to the manufacturer’s instruction. Briefly, 10μL of each related PCR product was mixed with 5 U of the corresponding restriction enzyme, 2μL of 10X buffers and 8μL H2O and then incubated 16h at 37 °C for BsmI and ApaI, 3h at 55 °C for FokI and 3 h at 65 °C for TaqI. Digested samples were resolved on 2.5% agarose gels as described above. Genotypes were scored blindly. Moreover, as a quality control, 10% of the samples were amplified and digested twice and second scored. Moreover, genotypes previously obtained in this sample at candidate genes consistently associated with the disease in different populations (HLA, INS, PTPN22, CTLA4, IL2RA, SUMO4, IFIH1, and CLEC16A) [18,19] were used, for the interaction tests. Genotyping was performed by polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) or tetra-primer amplification-refractory mutation system (ARMS)-PCR [20].
Autoantibodies evaluation
In this study, autoantibodies (AABs) for glutamic acid decarboxilase 65KDa (GAD65) and tyrosine phosphatase-like 2 (IA-2) were already tested for 100 cases [21,22]. For the present study, the remaining 100 samples were tested for the same two AABs, using a commercially ELISA-based Kit (AESKULISA, Mikroforum-Germany).
Data analysis
The study had over 80% power according to genetic power calculator [23]. Prior to association analysis, we performed QC of the obtained genotypes. Thus, data were tested for Mendelian inheritance using pedcheck [24]. Hardy-Weinberg equilibrium (HWE) for individual loci were assessed in parents using the Pearson chi-square test. Single-point and haplotype association analyses were carried out using two approaches: the transmission disequilibrium test (TDT) as implemented in Unphased [25] and conditional logistic regression as implemented in the “trio” R package [26]. A Bonferroni correction was applied for multiple testing, so p=0.05/12=0.004 (4 tests for individual SNPs plus 8 haplotypes).
Linkage disequilibrium (LD) was calculated and LD blocks were plotted using Haploview4.2 [27]. Haplotype analysis was performed using the 2- or 3- markers sliding windows method implemented in Unphased [23]. The sample was stratified according to the presence or absence of AABs and age at onset, as either early (≤5 years) or late (>5 years). For the gene-gene interaction, two-way gene-gene interaction of unlinked loci was tested using a conditional logistic regression as implemented in the “trio” R package [24].
Results
Sample description
This study included 200 nuclear families from Colombia. 105 out of 200 patients (52.5%) were male and the remaining 95 (47.5%) were females. Gender differences did not reach statistical significance ( p-value 0.33). The average age at diagnosis was 7.8 years (Table 1). Sixty one children (30.5%) developed the disease before 5 years age, with an average age for this category of 2.66 years. While the children with age at or above 5 yo had an average onset of 10.28 yo. Regarding the auto-antibodies, it was found that 78% (156) of the patients had at least one specific autoantibody related with T1D triggering (GAD-65 or IA-2), while 22% (44) were negative for these two AABs.
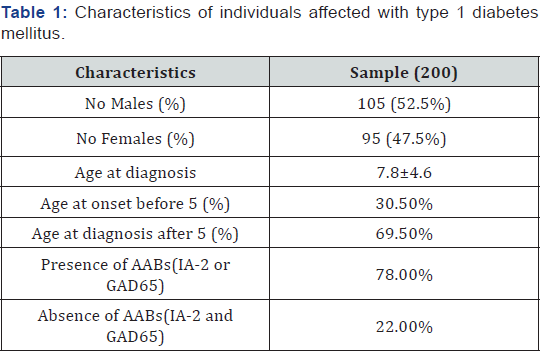
Unless indicated otherwise, data are given as the mean±SD or as the number of subjects in each group with percentages in parentheses.
Association and linkage disequilibrium analyses
Marker descriptions are given in Table 2. Data quality control showed that the genotyping rate was >98% and no Mendelian errors were found. The four SNPs were found to be in Hardy-Weinberg equilibrium. TDT single-point analysis did not detect any significant transmission distortion (Table 2). On the other hand, genotypic TDT based on 3 Pseudo controls (Table 3) revealed that genotype TT at SNP rs7675232 (ApaI) has a trend towards a protective effect (OR=0.59, 95% CI=0.34- 1.02; p=0.058). Not significant deviation of transmissions was observed when considering the presence or absence of AABs, or age at onset.
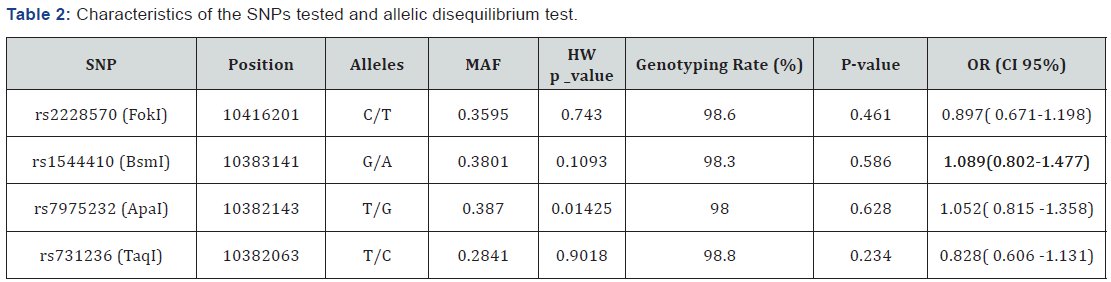
MAF: Minor Allele Frequeuncies; HE: Hardy-Weinberg Equilibrium; OR: Odds Ratio; CI: Confidence Interval
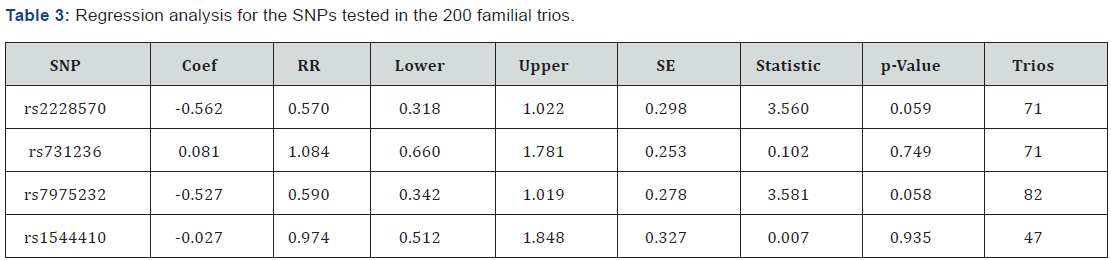
Results are for the recessive inheritance mode. Coef: Coeficiente; RR: Relative Risk; SE: Standard Error
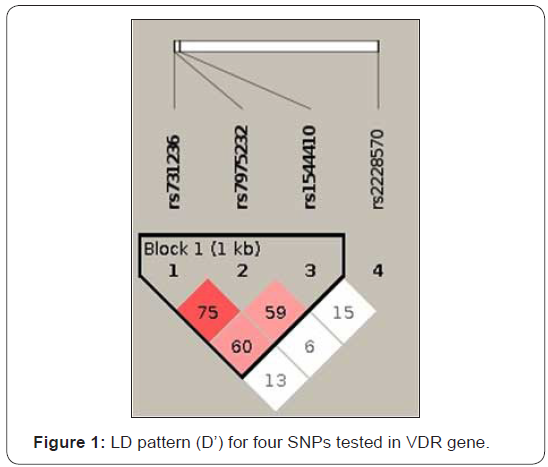
Linkage disequilibrium (LD) analysis revealed that 3 SNPs (rs1544410, rs7975232, rs731236) were in LD (Figure 1). Two haplotypes composed by alleles at these three loci were found to be associated with T1D in our sample (Table 4). These haplotypes were characterized by alleles A-G-T and G-T-C at at rs1544410-rs7975232-rs731236, respectively. Such haplotypeswere under-transmitted from parents to affected children (OR=0.219; ci95%=0.12-0.519 and OR=0.376; ci95% 0.18-0.786; p value=5.326x10-5). The most frequent haplotype was G-G-T (0.322), followed by haplotype G-T-T (0.235). A-G-T haplotype only reached 0.041 frequency. Haplotype stratified analysis according to the autoimmunity and age at onset revealed a significant associations (P-value=2.0x10-3). However, no significant confidence intervals were obtained (data not shown).
.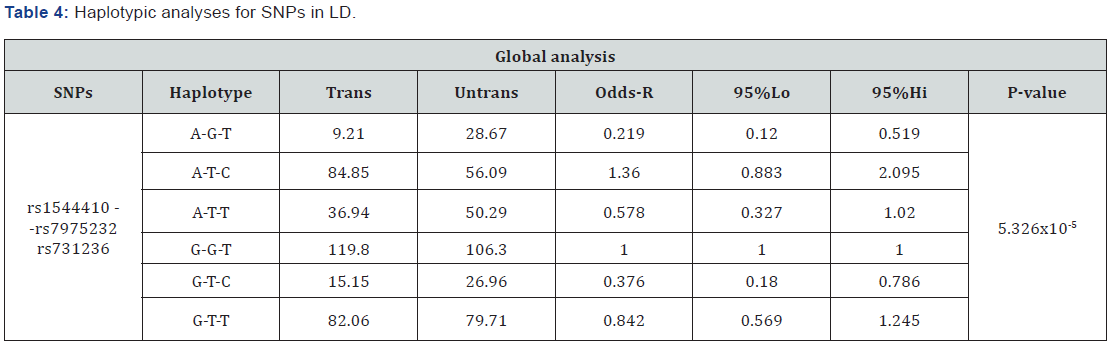
Trans: Transmissions; Untrans: Untransmissions
Bold values represent significant findings
Gene-gene interactions
Regarding the gene-gene interaction analysis, it showed a trend of interaction of VDR with CTLA4, IL2RA, HLA-I and HLA-II genes (Table 5). We identified G×G interactions between SNPs rs1544410 in VDR gene with rs3087243 in CTLA4 gene (p=8.9x10-3), rs7975232 in VDR and rs2040410 in HLA-II (p=0.01). In addition, SNP rs731236 in VDR appears to interact with SNP rs706778atIL2RA (p=0.01), and also interacts with SNPs rs7745906 and rs2524074 at HLA-I (p=0.01 and 0.02respectively). INS, PTPN22, SUMO4, IFIH1, and CLEC16A genes did not show significant interaction with VDR gene variants.
LL: Log Likelihood; IA: Interactions
Discussion
The purpose of this study was to test for association of VDR polymorphisms in a Colombian sample with T1D, all originated from Antioquia, Colombia. To our knowledge, this is the first study on the association between VDR SNPs and T1D among Colombian subjects. We examined the association of VDR gene polymorphisms at four positions (FokI, BsmI, TaqI and ApaI) with T1D. Associations between VDR polymorphisms and T1D have been seen in other countries including Hungary [17], Brazil [28], India [29], Croatia [30], Chile [31], Iran [12], Egypt [32], and other [4,33,34]. Meanwhile, other studies have not confirmed these findings and no associations were found neither in Portuguese [35] nor Danish populations [36], indicating that different genetic backgrounds may be causative of these discrepancies.
In the present study, we found that VDRApaI gene polymorphism (rs7975232) has a trend to protect in the recessive state. In agreement with our results, a meta-analysis found a significant association between ApaI polymorphism and T1D risk in East Asians [37]. We did not find any allelic association with T1D susceptibility. In fact, another recent metaanalysis showed that FokI, BsmI, ApaI and TaqI polymorphisms, when taken separately, seemed not to be associated with T1D risk, but particular haplotypes such as G-T-T and A-G-C (BsmIApaI- TaqI) showed significant associations [5]. None of these two haplotypes were found to be associated in our study. G-T-T haploytpe presented 0.235 frequency in our study (data not shown).
In addition, our results showed that haplotypes A-G-T and G-T-C protect significantly against T1D. Although different, others have also reported protective effects of specific vitamin D receptor (VDR) haplotypeS characterized by BsmI-ApaI-TaqI loci [38]. Even more, Uitterlinden et al. [11] found that haplotype A-G-T is one of the most frequent ones in Caucasians with a 50% prevalence. They also suggested that the variation of the degree of linkage disequilibrium between restriction fragment length polymorphism loci in the studied population might influence the degree of association between haplotypes and phenotypes. Our finding extends the repertoire of haplotypes associated with T1D, in different populations. This suggests a functional role of the VDR receptor in the susceptibility to T1D (perhaps extensive to autoimmunity as a whole), even in different haplotype contexts.
On the other hand, complex relationships may exist between VDR polymorphisms and other T1D related genes. Such interactions may explain why several previous findings regarding VDR polymorphisms have been inconsistent. In some cases, the effect might be missed if the gene is examined in isolation without allowing for its potential interactions with these other factors [39]. Our data showed a possible interaction effect between one of the VDR polymorphisms with CTLA4, IL2RA, HLA-I and HLA-IIloci.
It has been reported a biological interaction between vitamin D and CTLA4. CTLA4 is a critical suppressive protein that is expressed constitutively by regulatory T cells (Treg) and is induced on conventional T cells following activation. It functions to restrain inappropriate activation of autoreactive T cells and to restore T cell homeostasis following activation [40]. Jeffery et al. [41], found that vitamin D increases CTLA-4 expression through direct effects on the T cell, suggesting that the suppressive effect of vitamin D on T cell activation might be further enhanced through CTLA4-mediated removal of CD80/86, by transendocytosis. A possible explanation, why only SNP rs1544410 (BsmI) and not the complete haplotype in LD interacts with CTLA4, is that the LD presented corresponds to D’ and not to R2. R2 values are much smaller (data not shown).
VDR-1,25-(OH)2D3 complex has been shown to play a significant role in interfering with the signaling of transcription factors involved in the regulation of immunomodulatory genes[42] as well as expression of HLA class-II alleles on monocytes and human bone cells [43]. The interaction between VDR and HLA alleles may be mediated by the VDRE (VDR enhancer) present in the promoter region of HLA-DRB1*0301 allele [29]. Israni et al. [27] proposed that the insuficient amount of Vitamin D in early life in the predisposed individuals with HLA-DRB1*0301 can lead to an impaired expression of the allele in the thymus, resulting in escape from thymic deletion of autoreactive T cells. Furthermore, 1,25(OH)2D3 decreases MHC class I expression leading to reduced vulnerability of islet betacells to cytotoxic T lymphocyte [8].
On the other hand, it has been seen that IL2RA gene is regulated by vitamin D in CD4+ T cells. IL2RA is central to immuneregulation as an important modulator of immunity. IL2RA expression on regulatory T cells is essential for their function in suppressing T cell immune responses and autoimmune disease [44]. Exogenous addition of 1,25(OH)2D3 to CD4+ T cells in vitro induced the expression of IL2RA. This change in gene expression correlated with increased cell-surface expression of the IL2RA in activated CD4+ T cells [45]. For instance, in Multiple sclerosis patients, there was a significant correlation between IL2RA expression in CD4+ T cells and serum levels of 25 (OH)D3 [45].
This research has an important limitation, which regards to the sample size. It is clear that small sample sizes reduce the power to detect significant associations with less frequent variants or with variants with a small effect size on T1D. Also, all this study relies on the PCR-RFLP, which might not be the most reliable and thorough method, since other methods based on fluorescence could provide more reliable data. However, our study does have some strengths, such as the sample selection and the statistical approach (TDT). All of the individuals in this study belong to the most differentiated population in Colombia, which is a homogenous population living Northwest in the country. This population has reported sources of linkage disequilibrium, founder effects and ancestry characterization from its three-hybrid component, mainly European, followed by Amerindian and finally the African component [13-15,46,47]. Using the TDT, we have expected the associations not to reflect any false positive finding due to population stratification, since this only accounts the transmissions from heterozygous parents [48].
Conclusion
In conclusion, our study points to a protective role of the vitamin-D receptor gene in type 1 diabetes. It was found that two haplotypes at this gene (BsmI-ApaI-TaqI) protect significantly against T1D in our population. Furthermore, our results suggest an interaction between three VDR polymorphisms with HLA-II, HLA-I, IL2RA and CTLA4 SNPs, contributing to shed light on the plausible biological and biochemical pathways that underpin the disease. These suggestive interactions might be more clearly defined in a larger sample size than ours.
Acknowledgement
We are very grateful to the families who participated in this study. We are also very grateful to Dr Jorge Garcia-Ramírez for contributing patients. This study received financial support from Colciencias (grant contract # 0331-2013) and a doctoral scholarship (NG-L) from Colciencias call # 727. Also Universidad de Antioquia contributed to finance this study.
References
- Polychronakos C, Li Q (2011) Understanding type 1 diabetes through genetics: advances and prospects. Nat Rev Genet 12(11): 781-792.
- Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, et al. (2009) Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 41(6): 703-707.
- Kamel MM, Fouad SA, Salaheldin O, El razek AEAA, Abd AI (2014) Impact of vitamin D receptor gene polymorphisms in pathogenesis of Type-1 diabetes mellitus. Int J Clin Exp Med 7(12): 5505-5510.
- Wang G, Zhang Q, Xu N, Xu K, Wang J, et al. (2014) Associations between Two Polymorphisms (FokI and BsmI) of Vitamin D receptor gene and type 1 diabetes mellitus in Asian population: A Meta-Analysis. PLoS One 9(3): e89325.
- Tizaoui K, Kaabachi W, Hamzaoui A, Hamzaoui K (2014) Contribution of VDR polymorphisms to type 1 diabetes susceptibility: Systematic review of case-control studies and meta-analysis. J Steroid Biochem Mol Biol 143: 240-249.
- Chakhtoura M, Azar ST (2013) The role of vitamin d deficiency in the incidence, progression, and complications of type 1 diabetes mellitus. Int J Endocrinol 2013: 148673.
- Takiishi T, Van Belle T, Gysemans C, Mathieu C (2013) Effects of vitamin D on antigen-specific and non-antigen-specific immune modulation: relevance for type 1 diabetes. Pediatr Diabetes 14(2): 81-89.
- Riachy R, Vandewalle B, Belaich S, Kerr Conte J, Gmyr V, et al. (2001) Beneficial effect of 1,25 dihydroxyvitamin D3 on cytokine-treated human pancreatic islets. J Endocrinol 169(1): 161-168.
- Crofts LA, Hancock MS, Morrison NA, Eisman JA (1998) Multiple promoters direct the tissue-specific expression of novel N-terminal variant human vitamin D receptor gene transcripts. Proc Natl Acad Sci U S A 95(18): 10529-10534.
- Abd Allah SH, Pasha HF, Hagrass HA, Alghobashy AA (2014) Vitamin D status and vitamin D receptor gene polymorphisms and susceptibility to type 1 diabetes in Egyptian children. Gene 536(2): 430-434.
- Uitterlinden AG, Fang Y, Van Meurs JBJ, Pols HA, Van Leeuwen JP (2004) Genetics and biology of vitamin D receptor polymorphisms. Gene 338(2):143-156.
- Mohammadnejad Z, Ghanbari M, Ganjali R, Afshari JT, Heydarpour M, et al. (2012) Association between vitamin D receptor gene polymorphisms and type 1 diabetes mellitus in Iranian population. Mol Biol Rep 39(2): 831-837.
- Carvajal Carmona LG, Ophoff R, Service S, Hartiala J, Molina J, et al. (2003) Genetic demography of Antioquia (Colombia) and the Central Valley of Costa Rica. Hum Genet 112(5-6): 534-541.
- Carvajal Carmona LG, Soto ID, Pineda N, Ortíz Barrientos D, Duque C, et al. (2000) Strong Amerind/white sex bias and a possible Sephardic contribution among the founders of a population in northwest Colombia. Am J Hum Genet 67(5): 1287-1295.
- Pineda Trujillo N, Apergi M, Moreno S, Arias W, Lesage S, et al. (2006) A genetic cluster of early onset Parkinson’s disease in a Colombian population. Am J Med Genet B Neuropsychiatr Genet 141B(8): 885- 889.
- Howland JL (1995) Short protocols in molecular biology. In: John Wiley & Sons (Ed.), Short Protocols in Molecular Biology. (3rd edn), New York, USA, pp. 2-3.
- Györffy B, Vásárhelyi B, Krikovszky D, Madácsy L, Tordai A, et al. (2002) Gender-specific association of vitamin D receptor polymorphism combinations with type 1 diabetes mellitus. Eur J Endocrinol 147(6): 803-808.
- Rodríguez A, Alfaro JM, Balthazar V, Pineda Trujillo N (2015) Association analysis of PTPN22, CTLA4 and IFIH1 genes with type 1 diabetes in Colombian families. J Diabetes 7(3): 402-410.
- Rodriguez Rodriguez AM (2014) Evaluación de genes candidatos en una muestra de población colombiana con diabetes mellitus tipo.
- Ye S, Dhillon S, Ke X, Collins AR, Day IN (2001) An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res 29: E88.
- Pineda Trujilo N, Uribe F, Montoya F, Alfaro JM, Latorre G, et al. (2010) Chromosome region 2p25 is linked and associated with type 1 diabetes in Colombia. J Genet 89(4): 457-461.
- Gutiérrez achury J, Balthazar gonzález V, Bedoya berrío G, Ruíz linares A, Uribe londoño F, et al. (2009) Association of the TPO gene in Colombian families with type 1 diabetes. Iatreia 22(4): 323-329.
- Purcell S, Cherny SS, Sham PC (2001) Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19(1): 149-150.
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63(1): 259-266.
- Dudbridge F (2003) Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol 25(2): 115-121.
- Li Q, Fallin MD, Louis TA, Lasseter VK, McGrath JA, et al. (2010) Detection of SNP-SNP interactions in trios of parents with schizophrenic children. Genet Epidemiol 34(5): 396-406.
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2): 263- 265.
- Mory DB, Rocco ER, Miranda WL, Kasamatsu T, Crispim F, et al. (2009) Prevalence of vitamin D receptor gene polymorphisms FokI and BsmI in Brazilian individuals with type 1 diabetes and their relation to betacell autoimmunity and to remaining beta-cell function. Hum Immunol 70(6): 447-451.
- Israni N, Goswami R, Kumar A, Rani R (2009) Interaction of Vitamin D receptor with HLA DRB1*0301 in Type 1 diabetes patients from North India. PLoS One 4(12): e8023.
- Boraska V, Škrabić V, Zeggini E, Groves CJ, Buljubašić M, et al. (2008) Family-based analysis of vitamin D receptor gene polymorphisms and type 1 diabetes in the population of South Croatia. J Hum Genet 53(3): 210-214.
- García D, Angel B, Carrasco E, Albala C, Santos JL, et al. (2007) VDR polymorphisms influence the immune response in type 1 diabetic children from Santiago, Chile. Diabetes Res Clin Pract 77(1): 134-140.
- El Kafoury AA, Haroun M, Embaby AM, Dawoods AS (2014) The association of polymorphic sites in some genes with type 1 diabetes mellitus in a sample of Egyptian children. Egyptian Journal of Medical Human Genetics 15(3): 265-272.
- Zhang J, Li W, Liu J, Wu W, Ouyang H, et al. (2012) Polymorphisms in the vitamin D receptor gene and type 1 diabetes mellitus risk: an update by meta-analysis. Mol Cell Endocrinol 355(1): 135-142.
- Panierakis C, Goulielmos G, Mamoulakis D, Petraki E, Papavasiliou E, et al. (2009) Vitamin D receptor gene polymorphisms and susceptibility to type 1 diabetes in Crete, Greece. Clin Immunol 133(2): 276-281.
- Lemos MC, Fagulha A, Coutinho E, Gomes L, Bastos M, et al. (2008) Lack of association of vitamin D receptor gene polymorphisms with susceptibility to type 1 diabetes mellitus in the Portuguese population. Hum Immunol 69(2): 134-138.
- Thorsen SU, Mortensen HB, Carstensen B, Fenger M, Thuesen BH, et al. (2014) No association between type 1 diabetes and genetic variation in vitamin D metabolism genes: a Danish study. Pediatr Diabetes 15(6): 416-421.
- Wang Q, Xi B, Reilly KH, Liu M, Fu M (2012) Quantitative assessment of the associations between four polymorphisms (FokI, ApaI, BsmI, TaqI) of vitamin D receptor gene and risk of diabetes mellitus. Mol Biol Rep 39(10): 9405-9414.
- Ramos Lopez E, Jansen T, Ivaskevicius V, Kahles H, Klepzig C, et al. (2006) Protection from type 1 diabetes by vitamin D receptor haplotypes. Ann N Y Acad Sci 1079: 327-334.
- Cordell HJ (2009) Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet 10(6): 392-404.
- Gerold KD, Zheng P, Rainbow DB, Zernecke A, Wicker LS, et al. (2011) The soluble CTLA-4 splice variant protects from type 1 diabetes and potentiates regulatory T-cell function. Diabetes 60(7): 1955-1963.
- Jeffery LE, Qureshi OS, Gardner D, Hou TZ, Briggs Z, et al. (2015) Vitamin D Antagonises the Suppressive Effect of Inflammatory Cytokines on CTLA-4 Expression and Regulatory Function. PLoS One 10(7): e0131539.
- Morán Auth Y, Penna Martinez M, Shoghi F, Ramos Lopez E, Badenhoop K (2013) Vitamin D status and gene transcription in immune cells. J Steroid Biochem Mol Biol 136: 83-85.
- Ramagopalan S V, Maugeri NJ, Handunnetthi L, Lincoln MR, Orton SM, et al. (2009) Expression of the multiple sclerosis-associated MHC Class II Allele HLA-DRB1*1501 Is Regulated by Vitamin D. PLoS Genet 5(2): e1000369.
- Ounissi Benkalha H, Polychronakos C (2008) The molecular genetics of type 1 diabetes: new genes and emerging mechanisms. Trends Mol Med 14(6): 268-275.
- Berge T, Leikfoss IS, Brorson IS, Bos SD, Page CM, et al. (2016) The multiple sclerosis susceptibility genes TAGAP and IL2RA are regulated by vitamin D in CD4+ T cells. Genes Immun 17(2): 118-127.
- Rojas W, Parra MV, Campo O, Caro MA, Lopera JG, et al. (2010) Genetic make up and structure of Colombian populations by means of uniparental and biparental DNA markers. Am J Phys Anthropol 143(1): 13-20.
- Rishishwar L, Conley AB, Wigington CH, Wang L, Valderrama Aguirre A, et al. (2015) Ancestry, admixture and fitness in Colombian genomes. Sci Rep 5: 12376.
- Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52(3): 506-516.






























