Preparation of a DNA (Hepatoblastoma-Derived Cell Line: HepG2) Crown Cell Line
Shoshi Inooka*
Japan Association of Science Specialists
Submission:September 30, 2023;Published:October 11, 2023
*Corresponding author:Shoshi Inooka, Japan Association of Science Specialists, Japan
How to cite this article:Shoshi I. Preparation of a DNA (Hepatoblastoma-Derived Cell Line: HepG2) Crown Cell Line. J Tumor Med Prev. 2023; 4(2): 555635.DOI: 10.19080/ JTMP.2023.04.555635
Abstract
DNA crown cells (artificial cells), in which the outside of the membrane is covered with DNA, can be readily synthesized in vitro using sphingosine (Sph)-DNA-adenosine mixtures. These DNA crown cells can proliferate within egg whites in vivo. A previous report on the culture and synthesis of synthetic DNA (E. coli) crown cells showed that such a cell line could be prepared in approximately a month and maintained through four generations using synthetic DNA (Human placenta) crown cells. The present study examined whether a strain of synthetic DNA crown cells could be prepared using an established protocol using synthetic DNA (Hepatoblastoma-derived cell line; HepG2) crown cells. In addition, the microscopic characteristics of the prepared DNA (HepG2) crown cells are described.
Keywords:Synthetic DNA (Hepg2) Crown Cells; Cell Strain; Sphingosine-DNA; Cell Culture
Introduction
Artificial cells are cells that are covered with DNA, and they are referred to as DNA crown cells [1-3]. Synthetic DNA crown cells can be prepared and cultured using sphingosine (Sph)-DNA and adenosine-monolaurin (A-M) compounds, with incubation in egg white, respectively. In a series of studies on the cultivation of these cells in vitro, it has been shown that synthetic DNA (E. coli) crown cells could be cultivated for more than 50 days through seven generations, which qualified the cultured cells as a strain [4]. Moreover, it was shown that such a strain could be prepared in a shorter culture period and the protocol was presented [5,6]. Using this protocol, a strain of cells was prepared using synthetic DNA (Hepatoblastoma-derived cell line; HepG2) [7] crown cells cultured for three generations. The cultured cells were characterized microscopically.
Materials and Methods
Materials
The materials used were the same as those employed in a previous study [4]: Sph (Tokyo Kasei, Japan), DNA (from HepG2 cells), adenosine (Sigma-Aldrich; Wako, Japan), and monolaurin (Tokyo Kasei), adenosine-monolaurin (A-M) (a compound synthesized from a mixture of adenosine and monolaurin) [8,9]. Monolaurin solutions were prepared to a final concentration of 0.1 M in distilled water. Eggs, obtained from a local market, were used to obtain egg white. Dulbecco’s minimum essential medium (DMEM, Sigma-Aldrich) containing 10% bovine serum (Sigma-Aldrich) was used.
Methods
Preparation of synthetic DNA (HepG2) crown cells
Synthetic DNA (HepG2) crown cells were prepared as described previously [4]. Briefly, 180 μL of Sph (10 mM) and 90 μL of DNA (0.3oμg/μL) were combined, and the mixture was heated and cooled twice. A-M solution (100 μL) was added and the mixture was incubated at 37°C for 15 min. Next, 30 μL of monolaurin solution was added and the mixture was incubated at 37°C for another 5 min. The resulting suspension was used as the synthetic DNA (HepG2) crown cells.
Cell culture procedures
i.Two micro tubes were prepared and a total of 20 μL of sample was added to 200 μL of egg white. The mixture was then incubated for 7 days at 37oC (primary culture).
ii.Then, 50 μL of sample was added to 500 μL of DMEM and the mixture was incubated for 7 days at 37oC (secondary culture).
iii. Finally, 50 μL of sample was added to 500 μL of DMEM
and the mixture was incubated for a further 7 days at 37oC (third
culture; final sample).
The final samples were stored at approximately 4oC.
Microscopic observations
A total of 20 μL of the final samples were placed on a slide glass, covered with a cover glass, and observed under a light microscope.
Results and Discussion
i. Microscopic appearance of synthetic DNA (HepG2) crown cells in culture samples
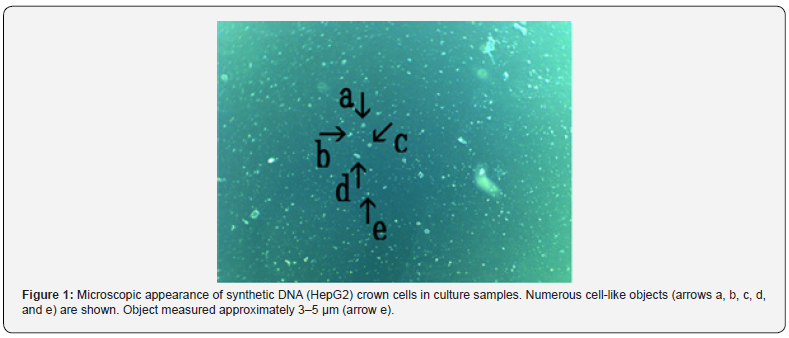
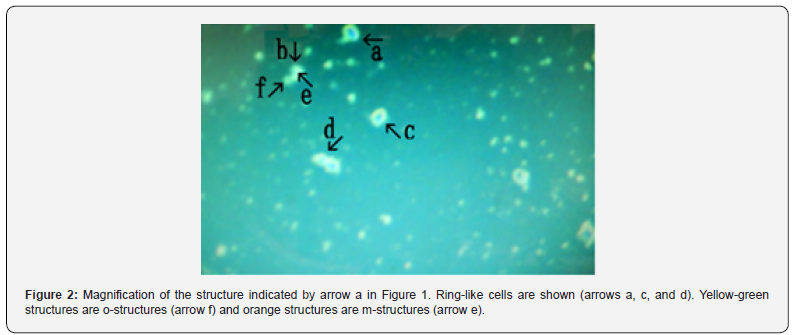
Figure 1 shows numerous cell-like objects (arrows a, b, c, d, and e). The object (arrow e) measured approximately 3–5 μm. Figure 2 shows a magnification of the structure indicated by arrow a in Figure 1.
Ring-like cells were observed (Figure 2 arrows a, c, and d). The different colors, which indicate different substances, facilitate explanations of other figures. For convenience, yellow-green structures are called o-structures (Figure 2 arrow f) and orange structures are called m-structures (Figure 2 arrow e), as described previously [10].
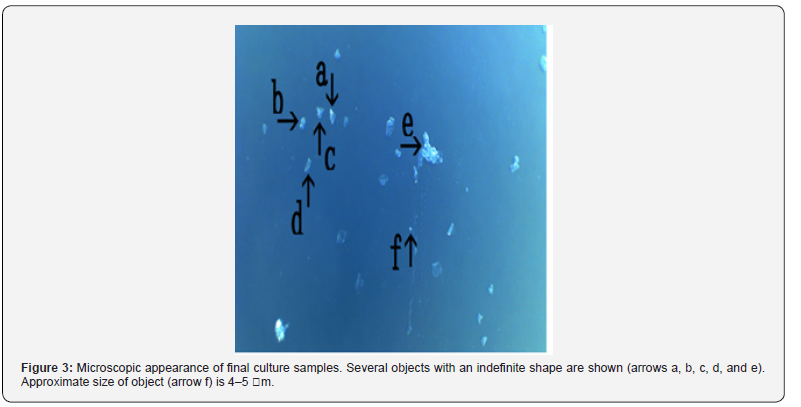
ii. Microscopic appearance of final cultures
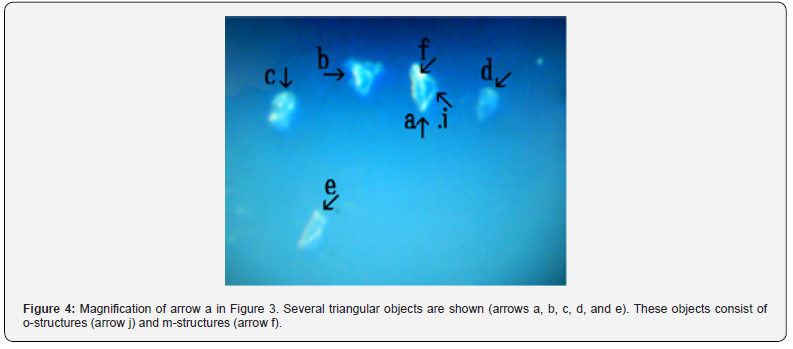
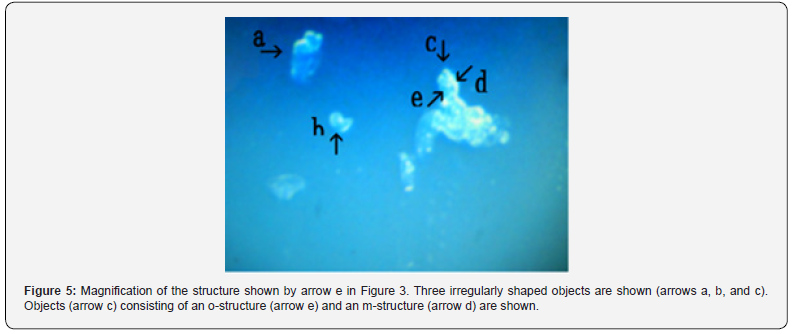
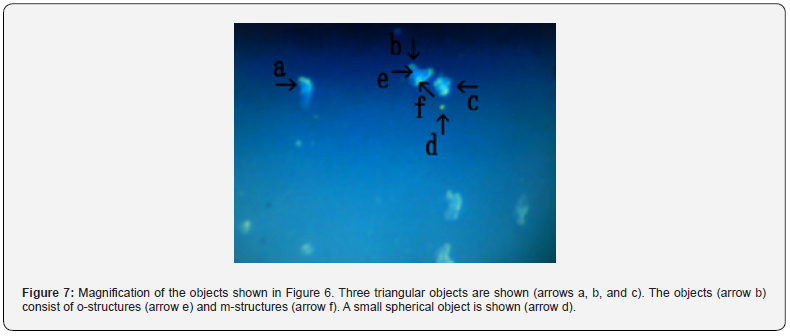
Figure 3 shows several objects with indefinite shape (arrows a, b, c, d, and e). The object (arrow f) measured about 4-5 μm. Figure 4 shows a magnification of the structure shown by arrow a in Figure 3. Several irregular triangular objects were observed (arrows a, b, c, d, and e). The objects consisted of o-structures (arrow j) and m-structures (arrow f) within o-structures. Interestingly, the shape of m-structures differed among objects (arrows a, b, and c). Figure 5 shows a magnification of the structure shown by arrow e in Figure 3. Three irregularly shaped objects of different sizes were observed (arrows a, b, and c). Also, an object (arrow c) consisting of an o-structure (arrow e) and an m-structure (arrow d) was observed. The m-structure (arrow d) in the object (arrow c) was elongated, whereas those in the objects indicated by arrows a and arrow b were rod-shaped or shaped like dot or rod. All objects were covered by o-structures (arrow e). Figure 6 shows the microscopic appearance of two objects in a final sample (arrows a and b). The approximate size of the object (arrow c) was 2-3 μm. Figure 7 shows a magnification of the structures shown in Figure 6. Three triangular objects were observed (arrows a, b, and c). The objects consist of o-structures (arrow e) and m-structures (arrow f). O-structures are gel-like, whereas m-structures are solid. Small spherical object which may be a cell were also observed (arrow d).

In previous experiments, synthetic DNA (E. coli) crown cells were cultured for seven generations over approximately 50 days [4]. In addition, in previous experiments with DNA (Human placenta) crown cells, cells were first cultivated in egg white and DMEM. In this way, a strain of cells was/could potentially be obtained within 30 days of culture [5]. Moreover, in the cultures of DNA (Akoya pearl oyster) crown cells, a strain could be prepared within approximately 21 days [6]. In the present experiments, a strain of DNA (HepG2) crown cells was prepared according to the method used to prepare DNA (Human placenta and Akoya pearl oyster) crown cells [5,6]. Although studies on the culture medium were not conducted, it is important that egg white is used as the medium for the first culture because synthetic DNA crown cells have been shown to multiply in egg white in vivo [2]. However, it is unclear whether the present culture medium is optimal for culturing synthetic DNA crown cells.
In contrast, it was unclear how the cultured synthetic DNA crown cells multiply and develop. During the cultivation of synthetic DNA crown cells, various kinds of objects were observed [4,10,11]. Hence, it is difficult to describe which cellular components are shared among them. On the other hand, round cells were observed in the prepared cells before culturing (Figure 2, arrows a, c, and d). Therefore, the basic cell shape may be round. However, in the present experiments, round cells were not observed and most cells were triangular. On the other hand, triangular cells have not been observed in the cultivation of synthetic DNA (E. coli, Human placenta and Akoya pearl oyster) crown cells [4, 5, 6]. However, it was not clear whether triangular shape was characteristic to HepG2 tumor cells. Most of the cultured cells or objects consist of o-structures (yellow-green color) and m-structures (orange color). O-structures were observed in the outside of cells, whereas m-structures were observed to be connected together inside o-structures.
On the contrary, in cultured HepG2 cells, o-structures seemed to have a gel-like consistency and diffuse (Figure 4, arrow j), whereas m-structures seemed to form solid like in various shape (Figure 4, arrow f).
The findings suggested that Sph-DNA-related components which had a gel-like consistency, were formed and covered the entire cells, because o-structures were Sph-DNA-related components. Recently, the multiplication of cultured synthetic DNA crown cells was demonstrated and it was shown that Sph- DNA-related components of synthetic DNA crown cells were elongated in the presence of adenosine-DNA [12]. These findings suggested that synthetic DNA crown cells were successfully cultivated. However, it is unclear whether adenosine-DNA form Sph-DNA related components which had a gel-like consistency. On the other hand, it was first time that synthetic DNA crown cells were prepared using DNA from tumor cells, and triangular cells were cultivated. To clarify whether such triangular cells are characteristic to tumor cells, additional synthetic DNA (tumor) crown cell strains need to be used, as these could potentially play a role in cancer research.
Acknowledgments
The author would like thank L. Monna (Rizo Inc., Tsukuba, Japan) for assistance in extracting DNA and for useful discussions. HepG2 cell lines were provided by the RIKEN BRC through the National BioResource Project of MEXT/AMED, Japan.
References
- Inooka S (2012) Preparation and cultivation of artificial cells. App Cell Biol 25: 13-18.
- Inooka S (2016) Preparation of Artificial Cells Using Eggs with Sphingosine-DNA. J Chem Eng Process Technol 7(1): 277.
- Inooka S (2016) Aggregation of sphingosine-DNA and cell construction using components from egg white. Integrative Molecular Medicine 3(6): 1-5.
- Inooka S (2022) Preparation of a DNA (E. coli) Crown Cell line in Vitro-Microscopic Appearance of Cells. Annals of Reviews and Research 8(1).
- Inooka S (2023) Preparation and Microscopic Appearance of a DNA (Human Placenta) Crown Cell Line. Journal of Biotechnology & Bioresearch 5(1).
- Inooka S (2023) Preparation of a DNA (Akoya pearl oyster) crown cell line. Applied Cell Biology Japan, p. 36.
- Doiores Lopez-Terada, Sau Wai C, Milton J Finegold, Barbara B Knowles (2009) HepG2 is a hepatoblastorma-derived cell line. Hum Pathol 40(10):1512-1515.
- Inooka S (2017) Biotechnical and Systematic Preparation of Artificial Cells. The Global Journal of Researches in Engineering 17: 1-10.
- Inooka S (2017) Systematic Preparation of Artificial Cells (DNA Crown Cells). J Chem Eng Process Technol 8(2): 327.
- Inooka S (2022) Microscopic appearance of synthetic DNA (E. coli) crown cells in primary culture. App Cell Biol Japan 35: 71-98.
- Inooka S (2022) Microscopic Appearance of Synthetic DNA (E. coli) Crown Cells in Secondary Cultures. Novel Research in Science 12(3).
- Inooka S (2023) Formation and Microscopic Appearance of Fireworks-like Objects Created from Synthetic DNA (E. coli) Crown Cells with Adenosine-DNA (E. coli) Annals of Review and Research 8(2).






























