Abstract
Background: Chronic rhinosinusitis with nasal polyps (CRSwNP) is a subtype of chronic rhinosinusitis that can be divided into two types: eosinophilic nasal polyps (ECRSwNP) and non-eosinophilic nasal polyps (NECRSwNP). These two types recur despite treatment with sinus medication and surgery, with ECRSwNP having a higher recurrence rate than NECRSwNP, and severely reduce patients’ quality of life while imposing a large economic burden. The pathogenesis and etiology of CRSwNP remain unclear and require further investigation.
Methods: In this study, differential gene expression analysis was performed on gene sequencing data obtained from the Gene Expression Omnibus (GEO) database. The key genes screened were verified using 14 pairs each of ECRSwNP and NECRSwNP clinical samples.
Results: Three of the screened genes (GPR97, NR4A3, and LIPN) were significantly overexpressed in ECRSwNP patients, suggesting that they play a critical role in the pathogenesis of ECRSwNP. Immune cell infiltration analysis of these genes showed that they were positively correlated with eosinophils and activated mast cells, and their expression was higher in nasal polyp tissues with high eosinophil and activated mast cell infiltration.
Conclusion: This study is the first to identify the key roles of GPR97, NR4A3, and LIPN in ECRSwNP and their relationship with immune cell infiltration. Our study provides new insights into the pathogenesis of ECRSwNP and suggests potential therapeutic targets.
Keywords: Chronic sinusitis with nasal polyps; Eosinophils; Transcriptome; Biomarkers; Mast cells.
Abbreviations:CDC: Conventional Dendritic Cells; SSC: Systemic Sclerosis; PCA: Prostate Cancer; TIF: Tubulointerstitial Fibrosis ; mPR3: Membrane Protease 3 ; MF : Molecular Functions ; CC: Cellular Components ; BP: Biological Processes ; DEGS: Differentially Expressed Genes; SD: Standard Deviation; GO : Gene Ontology ; KEGG: Kyoto Encyclopaedia of Genes and Genomes ; PPI: Protein–Protein Interaction; CRS : Chronic Rhinosinusitis; ARCI: Autosomal Recessive Congenital Ichthyosis ; NCBI: National Centre for Biotechnology Information
Introduction
Chronic Rhinosinusitis (CRS) is a highly heterogeneous rhino logic disease affecting the nasal and sinus mucosa that manifests in symptoms such as facial pain, a runny nose, congestion, and a decreased or lost sense of smell. The prevalence of CRS varies between countries and regions, with a prevalence rate of >10% in Europe and the United States and approximately 8% in China and Korea. Based on the presence or absence of polyps, CRS can be classified as either CRS without nasal polyps (CRSsNP) or CRS with nasal polyps (CRSwNP) [1-4]. Approximately 25–30% of patients with CRS have CRSwNP. CRSwNP can then be further subdivided into eosinophilic CRSwNP (ECRSwNP) and non-eosinophilic CRSwNP (NECRSwNP) based on the level of eosinophilic infiltration in the tissues, with ECRSwNP being predominantly associated with TH2 type inflammation and NECRSwNP being predominantly associated with TH1 and TH17 type inflammation [5,6]. CRSwNP is often exacerbated by aspirinexacerbated respiratory disease, allergic fungal rhinosinusitis, and asthma, and has a high rate of postoperative recurrence, with ECRSwNP having an especially high recurrence rate.
The pathogenesis of nasal polyps remains unclear, but most studies suggest that it is a complex multifactorial disease in which external environmental factors, including air pollutants, cigarette smoke, bacterial infections, epithelial damage, and inflammatory reactions, can play an important role [7-11]. Existing therapeutic drugs for treating nasal polyps are primarily corticosteroids and macrolide antibiotics. Corticosteroids are usually used to treat eosinophilic nasal polyps, but their long-term use can be harmful, and macrolide antibiotics are mainly used for the treatment of NECRSwNP [12]. Surgery is required when drug treatment is ineffective, but some patients have recurrence even after surgery, and thus require multiple surgeries, which seriously affects patients’ quality of life. Patients with eosinophilic nasal polyps usually have high eosinophilic infiltration and type 2 inflammation, with high tissue levels of Immunoglobulin E (IgE), interleukin-5 (IL-5), IL-4, and IL-13, as well as a significant increase in peripheral blood eosinophils. Patients with uncontrollable nasal polyps are mainly treated with omalizumab, dupleixumab, or mepolizumab [13-17]. Due to the high recurrence rate of the disease and the inability of current treatments to achieve a complete cure, nasal polyps have become a global health problem that seriously reduces the quality of human life and results in a large socioeconomic burden. Therefore, the pathogenesis of nasal polyps, and especially the pathogenesis of ECRSwNP, needs to be further explored.
In the present study, we screened the Differentially Expressed Genes (DEGs) in ECRSwNP compared with NECRSwNP and control samples. Using these genes, we performed Gene Ontology (GO) functional enrichment analysis and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment analysis, and we also analysed Protein–Protein Interaction (PPI) data. We identified potential key genes, thereby providing new insights into the pathogenesis of CRSwNP. Our study provides a foundation for the identification of candidate biomarker genes and drug targets.
Materials and Methods
Data sources
The National Centre for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih. gov/geo/) database stores many high-quality gene expression datasets. A high-throughput sequencing dataset GSE72713 was downloaded to classify ECRSwNP and NECRSwNP. submitted by Wang W et al. [18] and based on GPL11154 from the GEO database Illumina HiSeq 2000 (human) platform - including 3 eosinophilic nasal polyps (ECRSwNP), 3 non-eosinophilic nasal polyps (NECRSwNP) and 3 healthy control samples. Subjects with immunodeficiency disease, anterior anal polyps, fungal sinusitis, cystic fibrosis, primary ciliary dyskinesia, or gastroesophageal reflux disease were excluded from this study.
Screening for differential genes
Adapter sequences were removed from the raw sequencing data, and the individual libraries were converted to FASTQ format. Sequence reads were aligned to the human genome (hg19) using TopHat2, and the resulting sequence alignment files were reconstructed using Cufflinks and Scripture. For mRNA analysis, we chose the RefSeq database (Build 37.3) as the annotation reference. Transcripts with low confidence were filtered. Read counts for each transcript were normalized to the length of individual transcripts and the total mapped fragment counts in each sample and expressed as fragments per Kilobase Per Exon (FPKM) of mRNA per million fragments in each sample. Differential mRNA expression analysis was performed using Cuffdiff to compare groups: ECRSwNP vs. CTRL and ECRSwNP vs. non-ECRSwNP. corrected p values < 0.05 and | log2FC | > 1 were considered statistically significant.
GO and KEGG analysis
To explore the biological functions and pathways involved in significant co-DEGs and to understand the functional relationships between them, we performed Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) analyses on co-DEGs, where GO consists of Biological Process (BP), Cellular Component (CC) and Molecular Function (MF) components. GO and KEGG analyses were performed using the R package cluster Profiler, org.Hs.eg.db, and enrich plot, and P values < 0.05 were considered statistically significant. GO and KEGG enrichment results were plotted using an online website (https.//www. omicstudio.cn/tool).
Protein-protein interaction network analysis
Protein-protein interaction networks (PPI) were constructed using STRING (http://string-db.org) (version 11.5). PPI results were plotted using the software Cystoscope (version 3.9.1).
Clinical sample collection and qPCR experiment validation
A total of 28 patients who underwent surgical treatment at Run Hospital of Nanjing Medical University from 2020 to 2022 were recruited, including 14 ECRSwNP, 14 non-ECRSwNP uncinate process tissues and corresponding nasal polyp parietal tissues from nasal polyp patients (the clinical information of the patients is shown in Table 1), and the tissues were placed in lyophilized tubes after excision and stored in -80℃ refrigerator. For all enrolled patients, the diagnosis of ECRSwNP or non- ECRSwNP was made according to the EPOS 2020 [6]. E-CRSwNP was defined when the percentage of eosinophils in nasal mucosa exceeded 10% of the total infiltrating cells [19]. The study was approved by the Ethics Committee of Run Hospital of Nanjing Medical University (Ethical number: 2023-SR-017). Also, the patients were informed of the case sample collection.

Tissue RNA was extracted using Trizol reagent (Beijing Tiangen Biochemical Technology Co., Ltd.); cDNA was synthesized by reverse transcription using a PCR instrument (Hangzhou Langji Scientific Instruments Co., Ltd.) and a reverse transcription kit (ABM, G492). cDNA was synthesized using an ABI QuantStudio3 fluorescence quantitative PCR instrument (Applied Biosystems, Inc.) and a real-time fluorescence quantitative fluorescence kit (ABM, G891) for qPCR analysis. GAPDH gene expression was used as a standardized endogenous control. We used the ΔΔCt method to calculate relative gene expression. A set of primers was designed for these genes (as in Table 2).

Correlation analysis of target genes with immune cells
Immune cell infiltration analysis was performed on the CRSwNP dataset GSE72713 [20] using the online website CIBERSORTX (https://cibersortx.stanford.edu/), and the immune infiltration result data were correlated (Pearson) with the target gene expression from the sequencing results. Using online sites to create relevant heat mapping (https.//www.omicstudio.cn/tool).
Immunofluorescence triple-labelling of GPR97 or NR4A3, RNASE3, and TPSAB1
Nasal mucosal tissues from patients were fixed, embedded and made into paraffin sections, after deparaffinized and rehydrated, and the antigens were retrieved in boiling 10mM citrate buffer (PH6.0) for 8 min [21]. Then, they were incubated in 10% goat serum to block nonspecific antigens at room temperature for 30 min. Then, sections were incubated with antibodies against GPR97 (abmart, China, PA4659, 1:3000) RNASE3 (eosinophils marker, Proin tech, China, 55338-1-AP, 1:3000), TPSAB1 (mast cells marker, Proin tech, China, 13343-1-AP, 1:3000) or NR4A3 (abmart, China, TA0389, 1:3000), overnight at 4°C. On day 2, the sections were incubated with TSA Fluorescence System Kit including three different fluorescent dyes TYR-520, TYR-570, and TYR-690 sequentially for 10min. Then, they were incubated with DAPI solution for nuclear staining. Immunofluorescence images were acquired with a Digital Pathology Scanner.
Statistical analysis
Qualitative data are representative of at least three independent experiments. Quantitative data are presented as the means ± Standard Deviation (SD). Statistical differences between the two groups were determined using a two-tailed unpaired Student’s t-test. Statistical differences between multiple groups were compared using a one-way analysis of variance. Correlation analysis of the expression of two genes was assessed by Spearman’s rank test. All analyses were performed using GraphPad Prism 8.0 software (San Diego, USA). P <0.05 was considered statistically significant.
Results
Differential gene expression analysis with RNA-seq data
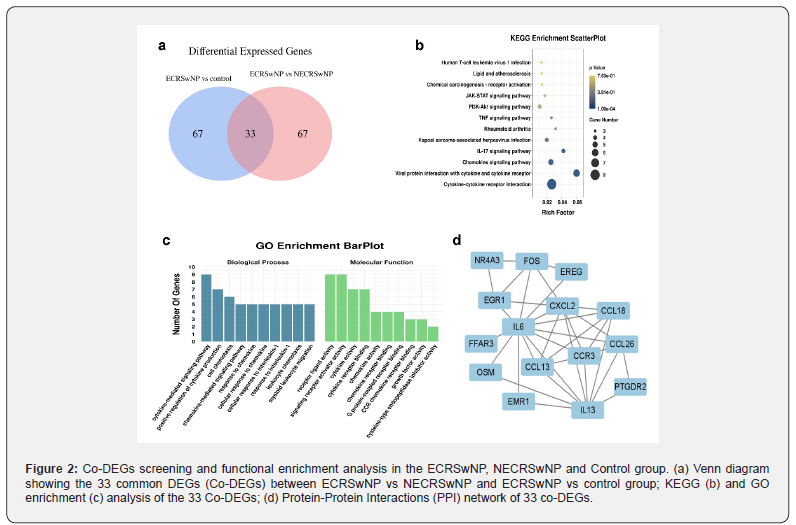
The flowchart of this study is shown in Figure 1a. First, the CRSwNP dataset in the GEO database was analysed for Differentially Expressed Genes (DEGs). Then, the top100 DEGs between the ECRSwNP group and the control group and ECRSwNP group and NECRSwNP group were selected. Taking the intersection, 33 common differential genes (co-DEGs) were found, and GO, KEGG, and PPI analyses were performed on these co-DEGs, A literature review was performed on these 33 co-DEGs, and 15 of them were found to have unstudied functions and mechanisms in nasal polyps. Subsequently, these 15 unstudied genes were validated using the clinical CRSwNP tissues.
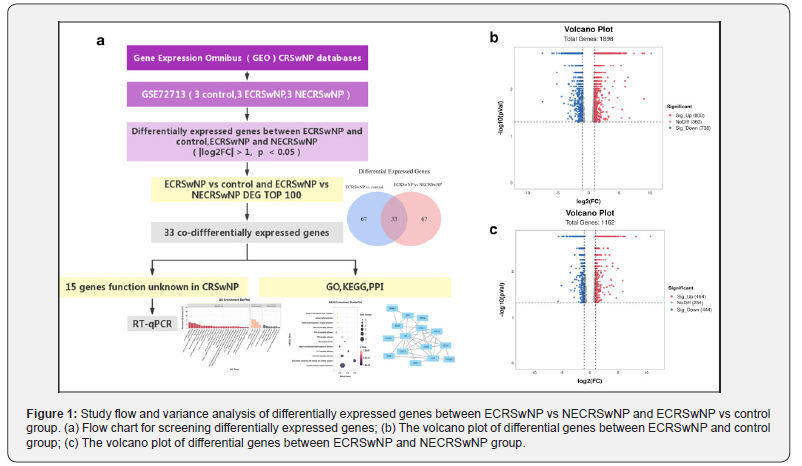
The high-throughput sequencing dataset GSE72713, which included three ECRSwNP samples, three NECRSwNP samples, and three normal control samples, was downloaded from the GEO database. After data preprocessing, DEG analysis was conducted. Between the ECRSwNP group and the control group, 1898 DEGs (adjust P <0.05 and |logFC| > 1) were found, of which 800 DEGs were significantly upregulated and 738 DEGs were significantly downregulated (as in Figure 1b); compared to NECRSwNP, 1162 DEGs were found in ECRSwNP DEGs (adjust P <0.05 and |logFC| > 1), of which 464 DEGs were significantly upregulated and 444 DEGs were significantly downregulated (as in Figure 1c).
Screening of key genes closely related to ECRSwNP occurrence and potential functional analysis
The top100 DEGs between the ECRSwNP group and the control group, and between the ECRSwNP group and the NECRSwNP group were analysed, and 33 co-DEGs were found (Figure 2a, Table 3). To explore the biological functions of these co-DEGs, we performed GO enrichment analysis using RStudio to investigate the Molecular Functions (MF), Cellular Components (CC) and Biological Processes (BP) most likely associated with differentially expressed genes. A total of 123 significantly enriched GO entries were found. 113 entries were enriched for the BP category, no entries were enriched for the CC category, and 10 entries were enriched for MF category. The most significant entries in the BP category were cytokine-mediated signaling pathways, and the most significant entries in the MF category were receptor ligand activity and signaling receptor activator activity Figure 2b). We also performed KEGG pathway enrichment analysis of the co-DEGs, highly enriched pathways included cytokine-cytokine receptor interaction, viral protein interaction with cytokine and cytokine receptor activity, viral protein interaction with cytokine, cytokine receptor activity, interaction with cytokine and cytokine receptors, and chemokine signaling pathway (Figure 2c). Furthermore, we constructed a PPI network using the STRING database to better understand the functional relationships between co-DEGs. The co-DEGs were converted into a PPI network, and Cystoscope was used to analyse and visualize the connectivity of each protein interaction sub-network (Figure 2d). These results suggest that these 33 co-DEGs may be closely related to the occurrence and development of nasal polyps.
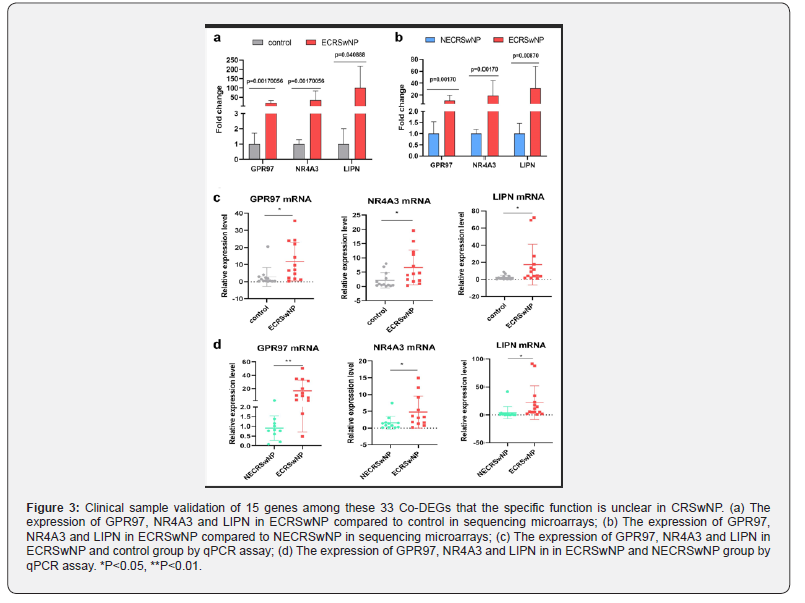
Clinical samples validation of 15 co-DEGs
A literature review revealed that among the above 33 co-DEGs, 15 genes had unknown functions in CRSwNP (Table 4), further validation of the relationship between these 15 genes and the development of CRSwNP is important for revealing the molecular mechanism of the disease. Next, we performed qPCR analysis on these genes in clinical samples of 14 pairs of eosinophilic polyps and 14 pairs of non-eosinophilic polyps clinical samples to validate the differential expression of these genes. The results showed that the expression of GPR97, NR4A3, and LIPN was significantly increased in nasal polyps of ECRSwNP patients compared with control tissues and NCRSwNP (Figure 3c, d), which was consistent with the sequencing results (Figure 3a, b). In conclusion, based on sequencing data and qPCR validation of clinical samples, we successfully identified three key genes (GPR97, NR4A3, and LIPN) associated with ECRSwNP, which may promote the formation of ECRSwNP.

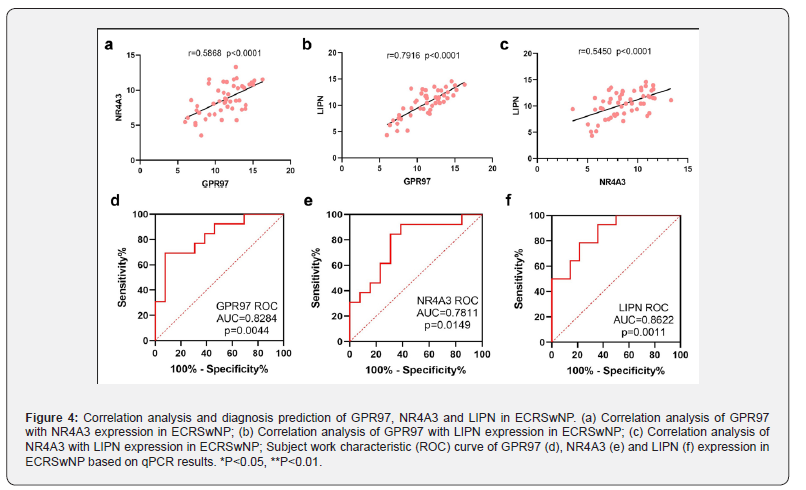
Correlation analysis and diagnostic value of GPR97, NR4A3, and LIPN
GPR97, NR4A3, and LIPN were highly expressed in ECRSwNP patients. Next, we analysed the correlations between these three genes in a pairwise manner. GPR97 was positively correlated with NR4A3, GPR97 with LIPN, and NR4A3 with LIPN (P < 0.0001), among which GPR97 was most significantly correlated with LIPN (Figure 4a-c). Regarding the quality of diagnostic prediction, the area under the ROC curve (AUC) values of GPR97, NR4A3, and LIPN were 0.8284, 0.7811, and 0.8622, respectively, according to the subject work characteristics (ROC) analysis, and the genes GPR97, NR4A3, and LIPN had a high diagnostic predictive value (AUC > 0.7) (Figure 4d-f). In addition, the GPR97, NR4A3, and LIPN levels related with clinical characteristics of the ECRSwNP patients were investigated. Correlation analysis results suggested that GPR97, NR4A3, and LIPN expression were positively correlated with counts of blood eosinophils in ECRSwNP patients, and LIPN levels was also significantly related with Sinus CT score (Table 5). Together, these findings suggest an important contribution of PR97, NR4A3, and LIPN to the CRSwNP diagnosis.


Functional analysis of GPR97, NR4A3, and LIPN
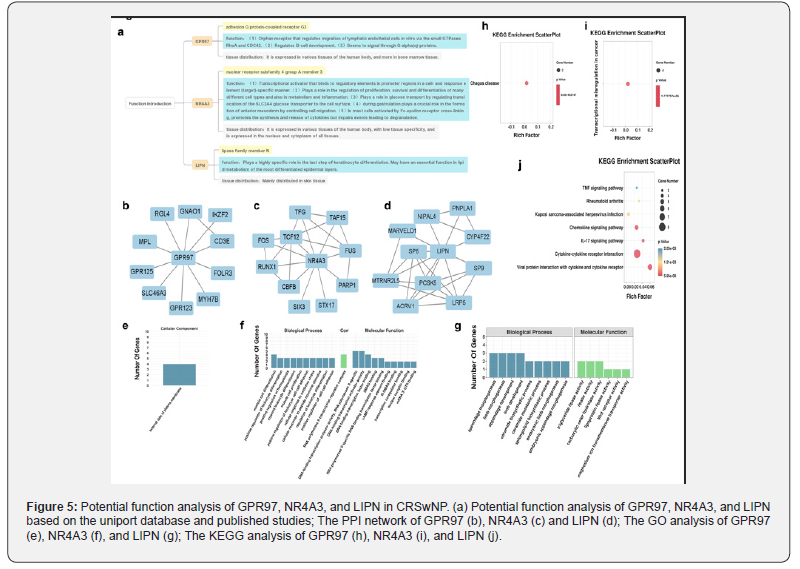
Since the functions and mechanisms of GPR97, NR4A3, and LIPN in CRSwNP are unknown, we first analysed the functions and distribution of these three genes in other diseases by UniProt database. We found that GPR97 regulates lymphatic endothelial cell migration and B-cell development; NR4A3 regulates cell proliferation, survival, and differentiation and in metabolism and inflammation; LIPN plays an important role in keratinocyte differentiation (Figure 5a). Next, we put these genes into the STRING database to identify their interacting proteins and conducted GO and KEGG pathway enrichment analyses (Figure 5b-d). For GPR97, we found one significantly enriched GO entry on the external side of the plasma membrane; for NR4A3 enrichment, we found 279 significantly enriched GO entries, including 247 entries in the BP category, 1 entry in the CC category, and 31 entries in the MF category; 15 significantly enriched GO entries were found for LIPN, including 9 entries in the BP category and 9 entries in the CC category (Figure 5e-g). KEGG pathway enrichment analysis showed that the most highly enriched pathway for GPR97 was Chagas disease; the most highly enriched pathway for NR4A3 was transcriptional mis regulation in cancer; and the most highly enriched pathway for LIPN was the Wnt signaling pathway (Figure 5h-j). Based on these results, we speculate that GPR97 plays a role in the migration of immune cells in nasal polyps, thus affecting polyp formation; NR4A3 may be involved in polyp formation through transcriptional processes, cell differentiation, and proliferation; and LIPN may be involved in the epithelial-mesenchymal transition process of nasal polyps through the Wnt and mTOR signaling pathways.
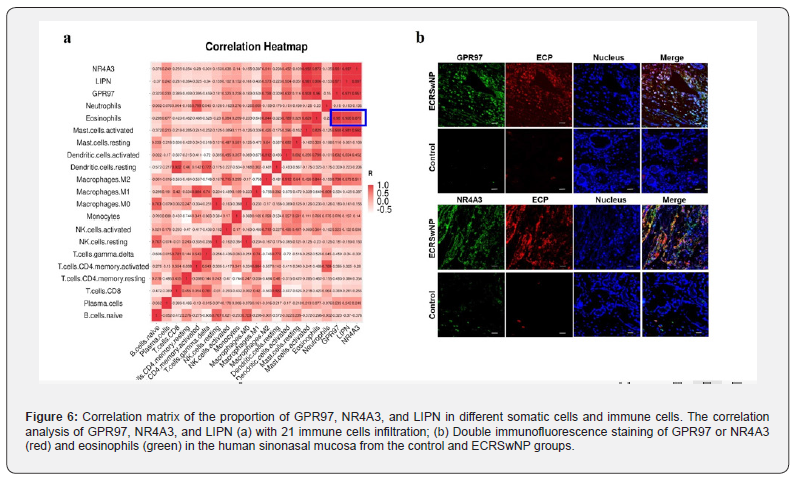
GPR97, NR4A3, and LIPN expression was positively correlated with eosinophils
To investigate the correlation between GPR97, NR4A3, and LIPN and immune cells, we applied the CIBERSORTX algorithm to calculate the relative abundance of 22 immune cells and integrated GPR97, NR4A3, and LIPN expression. The results showed that GPR97, NR4A3, and LIPN were highly positively correlated (r > 0.75) with eosinophils (Figure 6a). Since current commercially available LIPN antibodies do not meet the needs of tissue immunofluorescence, we examined the expression of GPR97 and NR4A3 in ECRSwNP and control tissues. Elevated expression of GPR97 and NR4A3 was found in ECRSwNP tissues, and co-localization with eosinophils (Figure 6b). This suggests that the increased expression of GPR97 and NR4A3 in ECRSwNP was positively correlated with eosinophils and may play an important role in promoting the occurrence of ECRSwNP.
Discussion
The formation of nasal polyps is closely linked to metabolic reactions, inflammatory responses, cytokines, immune responses, and environmental factors, and is a multistep, multifactorial, and complex process that involves alterations in the expression levels of multiple genes and their products, and multiple signaling pathways that regulate and antagonize each other, but the specific pathogenesis and etiology remain unclear. Therefore, identifying new biomarkers and understanding the underlying mechanisms are crucial for the treatment of CRSwNP.
Although the specific pathogenesis and etiology of CRSwNP are unclear, it is clear that the pathogenesis of ECRSwNP is different from that of NECRSwNP, with ECRSwNP being predominantly associated with T2 type inflammation, and NECRSwNP being predominantly associated with T1 and T3 type inflammation. Epithelial ILC2 and TH2 cells, and their interactions play a role in regulating the innate and adaptive immune responses in type 2 immunity in ECRSwNP [11]. Wang et al [22] performed scRNAseq on healthy individuals and heterogeneous CRS patients and found heterogeneity in ECRSwNP cells; for example, a unique type 2 immunity was determined by the cDC2-TH2 axis, ALOX15 macrophages (FN1 macrophages and FCER2 macrophages) were specifically enriched in ECRSwNP, and IL1B macrophages, CXCL9 macrophages and CXCL12 macrophages in NECRSwNP had higher ALOX5 expression than ALOX15 macrophages in ECRSwNP; in CRS, many of the epithelial cells remained undifferentiated basal cells, especially in ECRSwNP, and ECRSwNP patients exhibited a significant reduction in ciliated and secretory cells, suggesting that epithelial barrier function is impaired in CRS patients, with a higher degree of impairment in ECRSwNP patients. Our study revealed for the first time that GPR97, NR4A3, and LIPN are highly expressed in ECRSwNP, and these results combined with our immune cell infiltration analysis also showed that GPR97, NR4A3, and LIPN are highly positively correlated with eosinophils cells, suggesting that GPR97, NR4A3, and LIPN may play a key role in the pathogenesis of ECRSwNP.
GPR97, also known as ADGRG3 encodes adhesion G proteincoupled receptor G3, an adhesion-like G protein-coupled receptor. Ping et al [23] showed that the cryo-electron microscopic structure of the GPR97 Go complex bound to the anti-inflammatory drug beclomethasone or the steroid hormone cortisol and showed that the glucocorticoid partially bound within the transmembrane domain. Chu et al. [24] found that the adhesion molecule GPR97 activates CD177-related Membrane Protease 3 (mPR3) and binds to several proteins, leading to human neutrophil activation. GPR97 is a key factor in the progression of Tubulointerstitial Fibrosis (TIF) in patients with hypertension, and GPR97 promotes TGF-β signaling by facilitating transduction and promoting hypertension-associated TIF [25]. GPR97 was originally identified in mouse intestinal lymphatic endothelium and can regulate the migration of human lymphatic endothelial cells [26]. It is also highly expressed in some immune cells, such as mouse pre-B cells and thymocytes. It has also been shown that GPR97 has a role in regulating macrophage inflammation and metabolic dysfunction in high-fat diet-induced obese mice [27]. In high-fat diet-induced obese mice, deletion of GPR97 causes an increase in macrophages and accelerated invasion into the liver and kidney, suggesting that GPR97 may be involved in the development of local inflammation in obesity-associated renal tissues. Thus, high expression of GPR97 may contribute to ECRSwNP formation by similar mechanisms, leading to neutrophil activation and promoting TGF-β signaling.
NR4A3 (nuclear receptor subfamily 4 group A member 3) is a member of the NR4A family, which includes NR4A1-3. NR4A1, NR4A2, and NR4A3. These proteins drive immunosuppressive and fibrosis-related pathways in Conventional Dendritic Cells (CDC) in Systemic Sclerosis (SSC). Dysregulation of NR4A affects cytokine production in cDC2s and regulates downstream T cell activation [28]. NR4A3 maintains MT-2 signaling to inhibit Prostate Cancer (PCA) cell invasion, tumor growth, and metastasis, and supports tumor suppression by promoting p2 and PAI-21 expression as key factors in the TGF-β/Smad1 signaling pathway [29]. NR4A3 plays a key role in regulating the function of vascular cells, cardiomyocytes, and inflammatory cells as well as immune responses in cardiovascular remodeling in atherosclerosis, AAA, pulmonary hypertension, and cardiac hypertrophy [30]. NR4A3 plays a role in the migration of DC cells, which is required to bias monocyte differentiation toward monocyte-derived DCs at the expense of macrophages. NR4A3 is also able to regulate many genes in different cells and tissues and depending on the specific pathological physiological context and interactions with other regulatory pathways/transcription factors, the effects of NR4A3 on gene expression and cellular function vary across diseases. High NR4A3 expression in ECRSwNP may be associated with inflammatory responses and immune responses in mononuclear macrophages.
LIPN (lipase family member N), is a lipase that is highly expressed in granular keratin-forming cells of the epidermis and plays a role in the differentiation of these cells. Decreased expression of LIPN is associated with late Autosomal Recessive Congenital Ichthyosis (ARCI) and increases the expression of genes associated with ARCI and genes encoding proteins involved in lipid metabolism [31]. LIPN expression was found in systemic sclerosis (SSC), and LIPN is associated with amyl-tRNA biosynthesis, primary immunodeficiency, and DNA replication pathways [32]. cCAAT/enhancer binding protein α (C/EBPα) is a transcription factor that is abundantly expressed in the liver and white adipose tissue, and C/EBPα and the type 2 diabetic environment can lead to hepatic expression of LIPN [33]. In the past, little was known about LIPN, but it is now known that it is involved in keratinocyte differentiation and possibly also in the EMT process in nasal epithelial cells.
Although we have preliminarily confirmed that GPR97, NR4A3, and LIPN were highly expressed in ECRSwNP tissues, there are some limitations in the present study. First, in our study GPR97 and NR4A3 were positively correlated with high infiltration of eosinophils, but the specific role of GPR97 and NR4A3 in the formation of nasal polyps has not been clearly defined in vitro and murine model. Second, there are currently no LIPN antibodies to detect protein levels in samples, the importance role of LIPN can be detected at the genetic level by expanding clinical samples and constructing nasal polyps models and preparing LIPN antibodies to carry out related experiments in the future. Third, our study is the first to identify the relationship of GPR97, NR4A3, and LIPN in ECRSwNP, their regulatory mechanisms in the occurrence of nasal polyps need further research.
In summary, we identified three genes, GPR97, NR4A3, and LIPN, that are closely related to the pathogenesis of ECRSwNP. GPR97, NR4A3, and LIPN were significantly overexpressed in ECRSwNP, and their expression was highest in nasal polyps with high infiltration of eosinophils, which indicates that they play an important role in the pathogenesis of ECRSwNP. ROC curve analysis indicated that GPR97, NR4A3, and LIPN might be key regulators of ECRSwNP pathogenesis and serve as biomarkers. Our study provides insight into the pathogenesis of ECRSwNP. Further research on these new genes is of great significance for exploring the pathogenesis of ECRSwNP and searching for potential therapeutic targets.
References
- Bachert C, Marple B, Schlosser RJ, Hopkins C, Schleimer RP, et al. (2020) Adult chronic rhinosinusitis. Nat Rev Dis Primers 6(1): 86.
- Shi JB, Fu QL, Zhang H, Cheng L, Wang YJ, et al. (2015) Epidemiology of chronic rhinosinusitis: results from a cross-sectional survey in seven Chinese cities. Allergy 70(5): 533-539.
- Ahn JC, Kim JW, Lee CH, Rhee CS (2016) Prevalence and Risk Factors of Chronic Rhinosinusitis, Allergic Rhinitis, and Nasal Septal Deviation: Results of the Korean National Health and Nutrition Survey 2008-2012. JAMA Otolaryngol Head Neck Surg 142(2): 162-167.
- Sedaghat AR, Kuan EC, Scadding GK (2022) Epidemiology of Chronic Rhinosinusitis: Prevalence and Risk Factors. J Allergy Clin Immunol Pract 10(6): 1395-1403.
- Shi LL, Song J, Xiong P, Cao PP, Liao B, et al. (2014) Disease-specific T-helper cell polarizing function of lesional dendritic cells in different types of chronic rhinosinusitis with nasal polyps. Am J Respir Crit Care Med 190(6): 628-638.
- Fokkens WJ, Lund VJ, Hopkins C, Hellings PW, Kern R, et al. (2020) European Position Paper on Rhinosinusitis and Nasal Polyps 2020. Rhinology 58(Suppl S29): 1-464.
- Wang C, Yan B, Zhang L (2020) The epithelium-derived inflammatory mediators of chronic rhinosinusitis with nasal polyps. Expert Rev Clin Immunol 16: 293-310.
- Chegini Z, Didehdar M, Khoshbayan A, Karami J, Yousefimashouf M, et al. (2022) The role of Staphylococcus aureus enterotoxin B in chronic rhinosinusitis with nasal polyposis. Cell Commun Signal 20(1): 29.
- Zhong B, Seah JJ, Liu F, Ba L, Du J, et al. (2022) The role of hypoxia in the pathophysiology of chronic rhinosinusitis. Allergy 77(11): 3217-3232.
- Lee HJ, Kim DK (2022) Effect of Airborne Particulate Matter on the Immunologic Characteristics of Chronic Rhinosinusitis with Nasal Polyps. Int J Mol Sci 23(3): 1018.
- Kato A, Schleimer RP, Bleier BS (2022) Mechanisms and pathogenesis of chronic rhinosinusitis. J Allergy Clin Immunol 149(5): 1491-503.
- Kim DW, Yang SK (2022) Application of biologics in treating chronic rhinosinusitis with nasal polyps in Asian populations. Clin Exp Otorhinolaryngol 15(2): 125-126.
- Bachert C, Han JK, Wagenmann M, Hosemann W, Lee SE, et al. (2021) EUFOREA expert board meeting on uncontrolled severe chronic rhinosinusitis with nasal polyps (CRSwNP) and biologics: Definitions and management. J Allergy Clin Immunol 147(1): 29-36.
- Bachert C, Zhang N, Cavaliere C, Weiping W, Gevaert E, et al. (2020) Biologics for chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol 145(3): 725-739.
- Geng B, Bachert C, Busse WW, Gevaert P, Lee SE, et al. (2022) Respiratory Infections and Anti-Infective Medication Use From Phase 3 Dupilumab Respiratory Studies. J Allergy Clin Immunol Pract 10(3): 732-741.
- Fokkens WJ, Mullol J, Kennedy D, Philpott C, Seccia V, et al. (2023) Mepolizumab for chronic rhinosinusitis with nasal polyps (SYNAPSE): In-depth sinus surgery analysis. Allergy 78(3): 812-821.
- Gevaert P, Saenz R, Corren J, Han JK, Mullol J, et al. (2022) Long-term efficacy and safety of omalizumab for nasal polyposis in an open-label extension study. J Allergy Clin Immunol 149(3): 957-965.e3.
- Wang W, Gao Z, Wang H, Li T, He W, et al. (2016) Transcriptome analysis reveals distinct gene expression profiles in eosinophilic and noneosinophilic chronic rhinosinusitis with nasal polyps. Sci Rep 6: 26604.
- Cao PP, Li HB, Wang BF, Wang SB, You XJ, et al. (2009) Distinct immunopathologic characteristics of various types of chronic rhinosinusitis in adult Chinese. J Allergy Clin Immunol 124(3): 478-484 e1-2.
- Xiong G, Xie X, Wang Q, Zhang Y, Ge Y, et al. (2020) Immune cell infiltration and related core genes expression characteristics in eosinophilic and non‑eosinophilic chronic rhinosinusitis with nasal polyps. Exp Ther Med 20(6): 180.
- Qin D, Liu P, Zhou H, Jin J, Gong W, et al. (2022) TIM-4 in macrophages contributes to nasal polyp formation through the TGF-β1-mediated epithelial to mesenchymal transition in nasal epithelial cells. Front Immunol 13: 941608.
- Wang W, Xu Y, Wang L, Zhu Z, Aodeng S, et al. (2022) Single-cell profiling identifies mechanisms of inflammatory heterogeneity in chronic rhinosinusitis. Nat Immunol 23(10): 1484-1494.
- Ping YQ, Mao C, Xiao P, Zhao RJ, Jiang Y, et al. (2021) Structures of the glucocorticoid-bound adhesion receptor GPR97-Go complex. Nature 589(7843): 620-626.
- Chu TY, Zheng GC, Huang KY, Chang YC, Chen YW, et al. (2022) GPR97 triggers inflammatory processes in human neutrophils via a macromolecular complex upstream of PAR2 activation. Nat Commun 13(1):6385.
- Wu JC, Wang XJ, Zhu JH, Huang XY, Liu M, et al. (2023) GPR97 deficiency ameliorates renal interstitial fibrosis in mouse hypertensive nephropathy. Acta Pharmacol Sin 44(6):1206-16.
- Valtcheva N, Primorac A, Jurisic G, Hollmén M, Detmar M (2013) The orphan adhesion G protein-coupled receptor GPR97 regulates migration of lymphatic endothelial cells via the small GTPases RhoA and Cdc42. J Biol Chem 288(50):35736-35748.
- Shi J, Zhang X, Wang S, Wang J, Du B, et al. (2016) Gpr97 is dispensable for metabolic syndrome but is involved in macrophage inflammation in high-fat diet-induced obesity in mice. Sci Rep 6: 24649.
- Servaas NH, Hiddingh S, Chouri E, Wichers CGK, Affandi AJ, et al. (2023) Nuclear Receptor Subfamily 4A Signaling as a Key Disease Pathway of CD1c+ Dendritic Cell Dysregulation in Systemic Sclerosis. Arthritis Rheumatol 75(2): 279-292.
- Lin HY, Ko CJ, Lo TY, Wu SR, Lan SW, et al. (2022) Matriptase-2/NR4A3 axis switches TGF-β action toward suppression of prostate cancer cell invasion, tumor growth, and metastasis. Oncogene 41(20): 2833-2845.
- Martínez GJ, Cañes L, Alonso J, Ballester SC, Rodríguez SA, et al. (2021) NR4A3: A Key Nuclear Receptor in Vascular Biology, Cardiovascular Remodeling, and Beyond. Int J Mol Sci 22(21): 11371.
- Israeli S, Khamaysi Z, Fuchs TD, Nousbeck J, Bergman R, et al. (2011) A mutation in LIPN, encoding epidermal lipase N, causes a late-onset form of autosomal-recessive congenital ichthyosis. Am J Hum Genet 88: 482-487.
- Luo J, Li D, Jiang L, Shi C, Duan L (2023) Identification of Tregs-Related Genes with Molecular Patterns in Patients with Systemic Sclerosis Related to ILD. Biomolecules 13(3): 535.
- Aibara D, Matsuo K, Matsusue K (2022) Lipase family member N is a novel target gene for CCAAT/enhancer-binding protein α in type 2 diabetic model mouse liver. Endocr J 69(5): 567-575.






























