Taxonomic and Genetic Diversities of Cyanotoxin Production
Jian Yuan*
Department of Veterinary Diagnostic and Production Animal Medicine, College of Veterinary Medicine, Iowa State University, Ames, USA
Submission: June 06, 2020;Published: January 07, 2021
*Corresponding author: Jian Yuan, Department of Veterinary Diagnostic and Production Animal Medicine, College of Veterinary Medicine, Iowa State University, Ames, USA
How to cite this article: Jian Y. Taxonomic and Genetic Diversities of Cyanotoxin Production. JOJ Wildl Biodivers. 2021: 3(2): 555612 DOI: 10.19080/JOJWB.2021.03.555612
Mini Review
Cyanobacteria, colloquially known as blue-green algae, are a group of photoautotrophic microorganisms which exist ubiquitously in global freshwater bodies. They can bring about a few detrimental impacts, notwithstanding their essential role of important primary producer in aquatic ecosystems. Aside from the ecological harms caused by Harmful Algal Blooms (HABs) formed by explosive proliferation of cyanobacteria ascribed to proper growth factor such as sufficient nutrients and illumination, cyanobacteria can produce numerous kinds of cyanotoxins which are lethal to animals and humans at relatively low doses. These toxins are usually excreted by toxic cyanobacterial cells and dispersed in natural water, and victims are poisoned by mistakenly consuming the contaminated water. Since their habitats are water sources for most animals, and their growth is greatly enhanced under the circumstance of global warming, harms caused by cyanobacteria have further spread to terrestrial life such as livestock, wildlife, and pets. Numerous public and scientific reports concerning the poisoning and death of animals by cyanotoxins are seen hitherto.
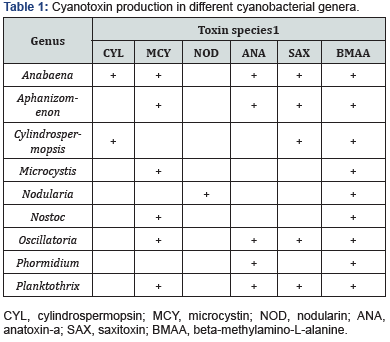
Cyanobacteria comprise a big deal of taxa, for example at the generic level, Anabaena, Cylindrospermopsis, Microcystis, Nodularia, Nostoc, Oscillatoria, Planktothrix, etc. On the other hand, cyanotoxins contain many species such as anatoxin-a, anatoxin-a(s), cylindrospermopsin, microcystin, and nodularin [1]. Although there are literal connections between some cyanobacterial genera and toxins, usually a toxin is not unique to a genus, and meanwhile, several toxins can be found in a genus (Table 1). This phenomenon complicates the diagnostic of culprit cyanobacterium when a cyanotoxin is tested, compared to the one-genus-one-toxin situation. Even though revelation of the presence, species, and level of cyanotoxin is necessary, confirmation of cyanobacterial producer(s) is still useful information for ecological investigation.
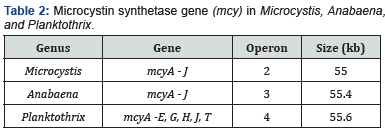
The genetic factor for cyanotoxin production is the presence of toxin synthetase genes which encode proteins responsible for enzymatic synthesis of toxins. The genes are clustered together so that they can be simultaneously expressed to make the proteins which are necessary for toxin synthesis [2]. As is introduced in the paragraph above, a toxin can be produced across a few genera except rare cases, for instance, nodularin’s exclusive production by Nodularia. Interestingly, sister species (i.e., in the same genus) producing the same toxin have identical toxin genes, while toxic intergeneric species have different genes with respect to gene compositions, operon loci, transcription directions, and sequences. For instance, microcystin can be found in Microcystis, Anabaena, and Planktothrix, but Microcystin Synthetase Genes (MCY) are distinct in each genus (Table 2). This further makes cyanotoxin production more diverse and complex at the genetic level. While toxigenicity can be determined by detection of the existence of toxin genes, it is difficult to cover all toxic cyanobacteria for a toxin because finding a common conserved domain across different generic gene clusters is challenging. Therefore, molecular detection of toxic cyanobacteria is usually conducted at the generic level, and multiplex PCR could be designed to compensate for being unable to establish an assay solely covering multiple toxic genera.
One of the possible reasons why the toxin synthetase genes of a certain cyanotoxin can be found in different genera is that Horizontal Gene Transfer (HGT) occurred in ancient cyanobacteria that have incorporated and reserved the genes till modern days [3]. However, these genes have experienced different mutational routes due to different evolution directions of relevant genera, resulting in the various extant forms of the clustered toxin genes. Because HGT is very popular and frequently occurring in bacterial microbes, it is most likely the toxin synthetase genes will be further diversified attributed to this rapid mutational phenomenon.
The taxonomic and genetic diversities of cyanotoxin production in cyanobacteria are no doubt one of the most complex natural toxigenesis phenomena. As a matter of fact, chemical modification of certain toxins makes more derivatives (e.g., over 100 microcystin variants) such that it is impossible to figure out a general principle for cyanotoxin production. However, as are increasingly disclosed the genetic fundament and synthetic mechanisms of more toxins, more efficient molecular diagnostic assays are anticipated for monitoring toxic cyanobacteria, and artificially engineered cyanobacteria may become a powerful ‘weapon’ to cope with HABs [4,5].
References
- Classen DM, Schwartz KJ, Madson D, Ensley SM (2017) Microcystin toxicosis in nursery pigs. J Swine Health Prod. 25(4): 198-205.
- Puschner B, Galey FD, Johnson B, Dickie CW, Vondy M et al (1998) Blue-green algae toxicosis in cattle. J Am Vet Med Assoc. 213(11): 1605-1607.
- Tillett D, Dittmann E, Erhard M, Dohren HV, Borner T et al (2000) Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide–polyketide synthetase system. Chem Biol. 7(10): 753-764.
- Rouhiainen L, Vakkilainen T, Siemer BL, Buikema W, Haselkorn R et al. (2004) Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaenastrain 90. Appl Environ Microbiol. 70(2): 686-692.
- Christiansen G, Fastner J, Erhard M, Borner T, Dittmann E (2003) Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation. J Bacteriol. 185(2): 564-572.






























