Abstract
This study investigated the biodegradation of the pesticide malathion by soil fungi, focusing on Alternaria alstromeriae KRMY EMIC001 (GenBank: PQ764546). The fungus demonstrated high degradation efficiency, achieving 89.5% degradation at 30ppm malathion under optimized conditions (25°C, pH 6, 21 days). Efficiency remained robust (86.5%) even at higher concentrations (150ppm). The study also examined the impact of different nitrogen and carbon sources on the degradation process. Additionally, the level of phosphatase enzyme in Alternaria alstromeriae KRMY EMIC001 was measured in the presence of malathion, showing a gradual decrease as malathion concentrations declined, highlighting the enzyme's role in the biodegradation process.
Keywords: Biodegradation; Malathion; Pollution; Alternaria alstromeriae; Phosphatase enzyme
Introduction
Malathion (C₁₀H₁₉O₆PS₂), a broad-spectrum organophosphorus insecticide, inhibits acetylcholinesterase and is extensively used in agriculture and public health programs. Despite its efficacy, malathion and its degradation byproducts (e.g., malaoxon) exhibit heightened neurotoxicity and environmental persistence, threatening ecosystems and human health—particularly in densely treated regions [1]. In Egypt, its affordability has exacerbated contamination of soil and water systems [2], necessitating sustainable remediation strategies. Conventional malathion removal via chemical oxidation or adsorption is costly and generates secondary pollutants [3]. In contrast, microbial degradation leverages enzymatic pathways (e.g., phosphotriesterases) to mineralize malathion into non-toxic intermediates [4]. While bacterial degraders (e.g., Pseudomonas spp.) are well-characterized [5]. Pesticides, including malathion, represent a diverse group of chemical compounds with varying physicochemical properties, necessitating the development of specialized analytical methods and equipment. This diversity can pose challenges for many laboratories, particularly in resource-limited settings [6]. Human exposure to pesticides occurs primarily through ingestion, inhalation, or dermal contact, with ingestion being the most significant route of contamination, followed by inhalation [7,8]. Fungal systems—particularly soil-inhabiting genera like Alternaria—remain overlooked. Existing research prioritizes white-rot fungi (e.g., Phanerochaete chrysosporium), despite evidence that Alternaria spp. thrive in pesticide-contaminated soils and secrete hydrolytic enzymes (e.g., phosphatases) with untapped bioremediation potential. We report the isolation and optimization of Alternaria alstromeriae KRMY EMIC001 (GenBank: PQ764546), a newly documented fungal strain capable of degrading 89.5% of malathion (30ppm) within 21 days. Fungal systems offer distinct advantages over bacteria, including: Environmental resilience: Tolerance to pH/temperature fluctuations (optimal at pH 6, 25°C). Enzymatic efficiency: Extracellular phosphatase production correlates with degradation rates. Scalability: Cost-effective growth in low-nutrient conditions, ideal for resource-limited settings. This study bridges a critical gap in fungal bioremediation, positioning A. alstromeriae as a viable agent for large-scale malathion detoxification.
Materials and Methods
Collection of soil and water samples for fungal isolation
Soil and water samples were collected from various locations in Kafr El-Zayat City, Gharbia Governorate, Egypt. Soil samples were collected from pesticide-contaminated sites at a depth of 5-10cm, air-dried, and sieved through a 2mm mesh. The sieved samples were stored in sterile polyethylene bags and transported to the laboratory, where they were kept at 3°C until further processing. Water samples were collected in sterilized glass bottles and processed within 24 hours of collection [9].
Cultural medium
Czapek-Dox agar medium was used for the isolation, screening, and cultivation of fungi. The medium was prepared by dissolving the following components in 1000 mL of distilled water (g/L): sucrose (30.0), sodium nitrate (2.0), potassium dihydrogen phosphate (1.0), potassium chloride (0.5), magnesium sulfate (0.5), ferrous sulfate (0.01), and agar (15.0). The pH was adjusted to 7.3 ± 0.2, and chloramphenicol (0.05g/L) was added as a bacteriostatic agent. The medium was autoclaved at 121°C for 15 minutes before use [10].
Fungal isolation and identification
Fungi were isolated from soil and water samples using the serial dilution method [11]. Aliquots of the diluted samples were inoculated onto Czapek-Dox agar plates and incubated at 28 ± 2°C. Fungal colonies were subcultured onto fresh medium and purified using either the hyphal tip or single spore technique. Purified isolates were transferred to Czapek-Dox agar slants for further identification and characterization.
Inoculum preparation
Fungal inoculum was prepared by growing the isolates on sterilized potato dextrose agar (PDA) slants for 5 days at 30°C. Spore suspensions were prepared by adding 0.9% NaCl solution containing a few drops of Tween 80 to each slant and gently brushing the surface with an inoculating loop under aseptic conditions. The spore suspension was diluted, and clumps were dispersed by stirring. Serial dilutions were prepared in 0.1% Tween 80, and spore counts were determined using a hemocytometer. A spore suspension containing 1 × 10⁶ spores/mL was used as the inoculum [12].
Screening for pesticide degradation by isolated fungi
Screening on solid medium
The degradation capability of the fungal isolates was assessed using Czapek-Dox agar plates supplemented with malathion at concentrations of 0, 30, 60, 90, and 150ppm. Chloramphenicol (0.05% w/v) was added to prevent bacterial contamination, and the pH was adjusted to 7.3 using 1N HCl. Plates were inoculated with a 9mm fungal disc and incubated at 28°C for 7 days. Fungal growth and tolerance to malathion were evaluated by measuring colony diameter and comparing it to control plates without malathion.
Screening in liquid medium
screening was performed in 250mL Erlenmeyer flasks containing 100mL of Czapek-Dox broth supplemented with varying concentrations of malathion. The pH of the medium was adjusted to 7.0, and chloramphenicol was added after autoclaving. Each flask was inoculated with 1 × 10⁷ spores/mL and incubated in a rotary shaker at 28°C and 130rpm for 7 days. After incubation, the culture was filtered through Whatman No. 2 filter paper, and the fungal biomass was determined by dry weight measurement. The filtrate was centrifuged at 10,000rpm for 10 minutes, and the supernatant was collected for spectrophotometric analysis of malathion degradation.
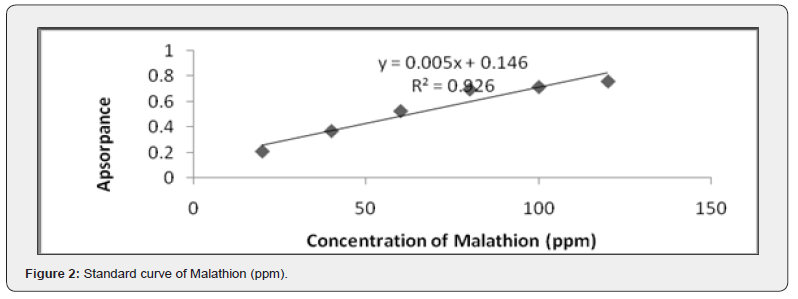
Determination of malathion degradation by spectrophotometry
The maximum absorbance wavelength for malathion was determined to be 520nm. A standard curve was constructed using known concentrations of malathion to quantify its degradation. The percentage of malathion degradation was calculated based on the reduction in absorbance at 520nm after fungal treatment (Figure 2).
Identification of the isolated fungus with high degradation ability (isolate no. 2)
Fungal isolate No. 2 demonstrated the highest malathion degradation capability and was selected for further characterization. Pure cultures of the isolate were prepared and identified using light microscopy. Morphological identification was based on colony characteristics, cultural features, and the structure of sporulating organs, following standard mycological keys [13,14].
Molecular identification of fungal strains
The fungal strain (isolate No. 2) was further identified by 18S r RNA sequence analysis. DNA extraction was performed using a modified Chelex protocol: 20µL Chelex, 40µL TE buffer, and fungal mycelia were heated to 95°C for 2 minutes, followed by centrifugation at 13,000 × g for 2 minutes. The supernatant containing DNA was transferred to clean tubes and stored at -18°C.
The internal transcribed spacer (ITS) region was amplified using PCR with the following conditions: 35 cycles of denaturation at 94°C for 35 seconds, annealing at 55°C for 55 seconds, and extension at 72°C for 45 seconds, with a final extension of 7 minutes at 72°C. The primers used were ITS1 (5’-TCCGTAGGTGAACCTGCGG-3’) and ITS4 (5’-TCCTCCGCTTATTGATATGC-3’) [15,16]. PCR products were sequenced by Macrogen (Animal Health Research Institute (AHRI), and sequences were aligned using MEGA version 5. A BLAST search in Gen Bank identified the most similar sequences. Phylogenetic analysis was performed using the Maximum Likelihood method in MEGA version 5, with bootstrap support based on 1,000 replicates. The isolate was identified as Alternaria alstromeriae KRMY EMIC001.
Optimization of Growth Conditions for Malathion Biodegradation by Selected Isolates.
The growth conditions for isolate No. 2 were optimized in Czapek-Dox liquid medium under varying physiological parameters:
pH: 2, 4, 6, and 8
Temperature: 20, 25, 27, and 30°C
Incubation time: 7, 14, 21, and 30 days
Carbon sources: Zero carbon, glucose, fructose, sucrose, maltose, lactose, and starch
Nitrogen sources: Zero nitrogen, sodium nitrate, ammonium nitrate, urea, asparagine, and peptone
Cultures were incubated under shaking conditions at 130rpm.
Assay of phosphatase activity
Phosphatase activity was measured alongside malathion degradation. Samples were centrifuged at 10,000× g for 15 minutes at 4°C, and fungal cells were washed three times with 10mmol L⁻¹ sodium carbonate/bicarbonate buffer (pH 10.5). Cells were resuspended in 5mL of the same buffer and sonicated for 10 seconds (10 cycles with 15-second intervals) to release phosphatase enzymes. The supernatant was collected after centrifugation to remove cell debris.
Phosphatase activity was determined using an alkaline phosphatase kit (Spectrum, Egypt). The working solution was prepared by mixing 900µL buffer (R1) with 100µL substrate (R2). A 10µL sample was added to 1mL of the working solution, and absorbance was measured at 405nm using a DTN-405 semi-automated chemistry analyzer. The mean absorbance change per minute (ΔA/min) was calculated [17].
Gas chromatography-mass spectrometry (GC-MS) analysis
After the incubation period, 100mL of growth medium containing malathion was mixed with 50mL dichloromethane and 40mL of 20% sodium chloride solution in a 500mL separatory funnel. The mixture was partitioned three times with dichloromethane, filtered through anhydrous sodium sulfate, and evaporated to dryness using a rotary evaporator at 30°C. The residue was dissolved in 1mL acetone, filtered through a 0.45µm syringe filter, and 200µL was injected into the GC-MS system.
GC-MS analysis was performed using a Trace GC Ultra-TSQ Quantum mass spectrometer (Thermo Scientific, USA) equipped with a TG-5MS capillary column (30m × 0.25mm × 0.25µm). The oven temperature was held at 70°C for 5 minutes, then increased to 280°C at 5°C/min and held for 5 minutes. The injector and MS transfer line temperatures were set at 250°C. Helium was used as the carrier gas at a flow rate of 1 =mL/min. The system operated in SIM/SCAN mode, with a solvent delay of 4 minutes [18].
Statistical analysis
All experiments were conducted in triplicate. Mean values and standard deviations were calculated using Microsoft Excel 2010.
Results and Discussion
Isolation and screening of malathion-degrading fungal strains
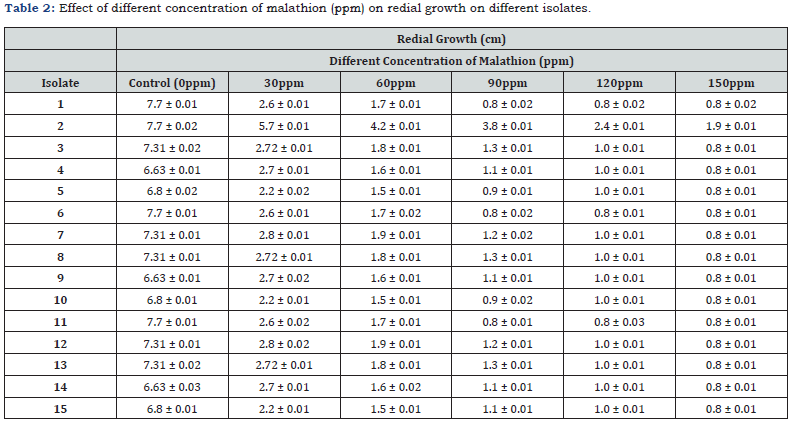
Fifteen fungal strains were isolated from soil samples and three from wastewater adjacent to Kafr El-Zayat Pesticides Company (KEC), Egypt (Table 1). Initial screening under varying malathion concentrations (30-150ppm) identified six isolates (1–6) capable of degrading the pesticide. Isolate 2 (Alternaria alstromeriae KRMY EMIC001) exhibited superior radial growth (7.7 ± 0.02cm in control; 1.9 ± 0.01cm at 150ppm) and degradation efficiency (Table 2), outperforming other strains. This aligns with reports of Alternaria spp.’s bioremediation potential [3] and enzymatic organophosphate degradation mechanisms observed in Fusarium oxysporum [4].
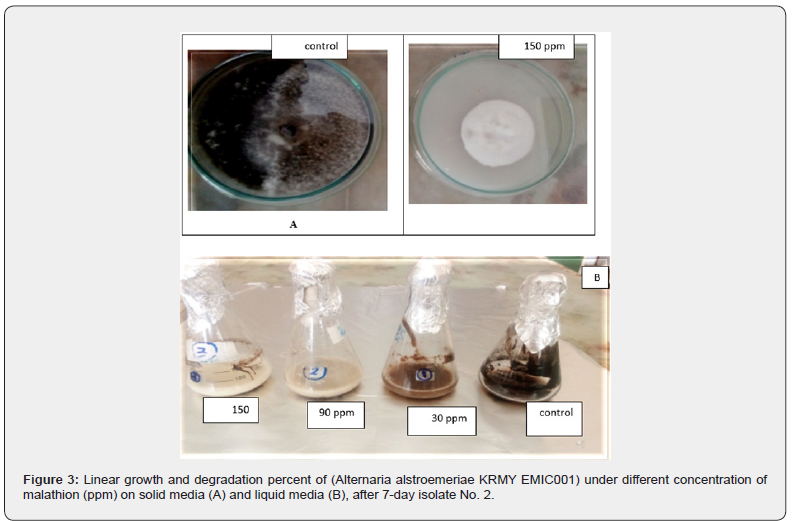
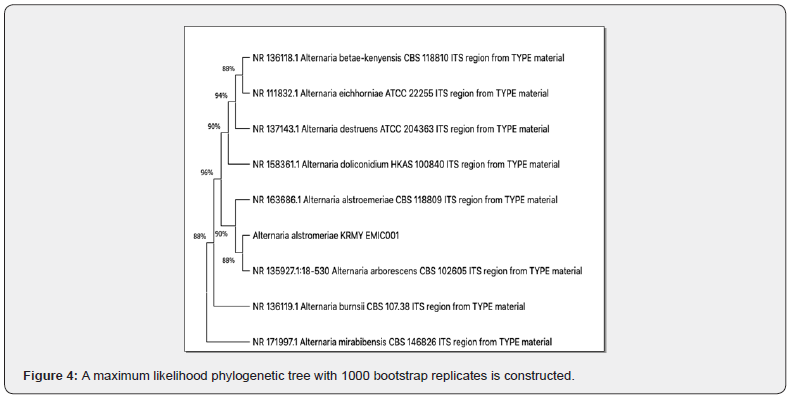
Molecular identification and phylogenetic analysis
ITS sequencing (1,391bp) of isolate 2 revealed 96% nucleotide identity with Alternaria alstromeriae KRMY EMIC001 (NCBI: PQ764546). Phylogenetic analysis (MEGA 5, Maximum Likelihood, 1,000 bootstrap replicates) confirmed its taxonomic position within the Alternaria clade (Figure 4), consistent with ITS-based fungal identification [19].
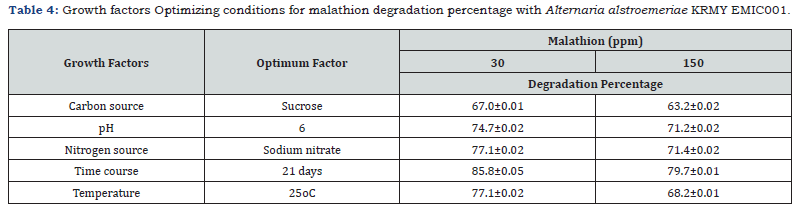
Optimization of malathion degradation
Optimal degradation by A. alstromeriae occurred at pH 6, 25°C, with sucrose (67.0 ± 0.01% at 30ppm) and sodium nitrate (77.1 ± 0.02% at 30ppm) as carbon and nitrogen sources, respectively (Table 4). Prolonged incubation (21 days) enhanced degradation to 85.8 ± 0.05% (30 ppm) and 79.7 ± 0.01% (150ppm), mirroring enzymatic adaptation kinetics in Pleurotus ostreatus [20]. The pH and temperature optima correlate with fungal phosphatase activity [21].

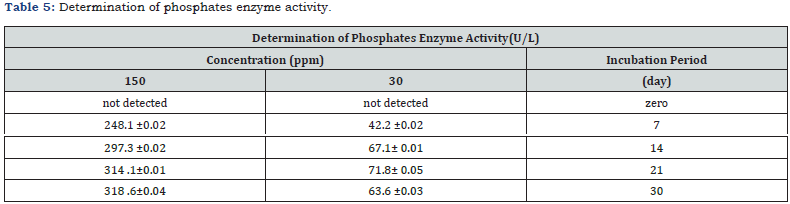
Phosphatase activity and enzymatic degradation
Phosphatase activity peaked at 318.6 ± 0.04U/L (150ppm, 30 days), correlating with malathion degradation (Table 5). This suggests phosphatase-mediated hydrolysis of P–O–C bonds, yielding less toxic metabolites (e.g., dimethyl thiophosphate), consistent with organophosphate degradation pathways [22].
GC-MS analysis of degradation products
GC-MS confirmed an 89.5% reduction in malathion after 21 days, with dimethyl 3-thiaadipate (13.962% abundance) as a primary metabolite (Figure 5, Tables 6-7). The absence of toxic intermediates (e.g., phosphorodithioic acid) and the prevalence of alkanes/esters indicate detoxification via P–S bond cleavage, a mechanism observed in Fusarium spp. [2,23].
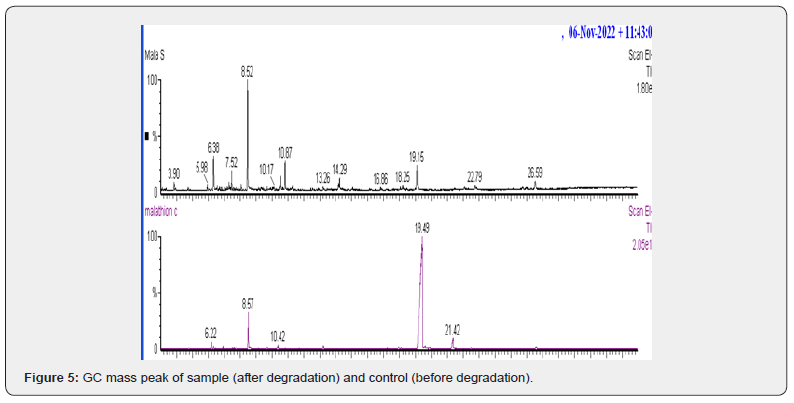
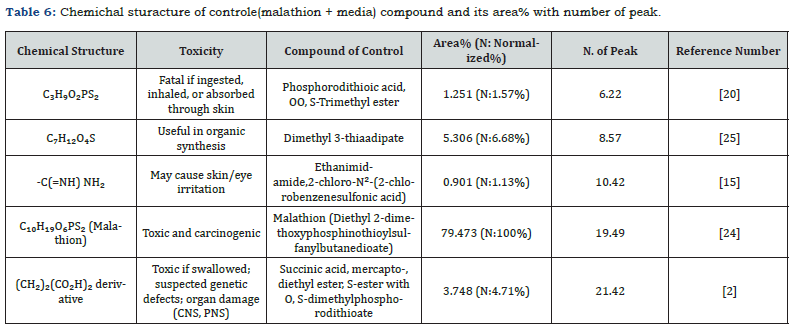
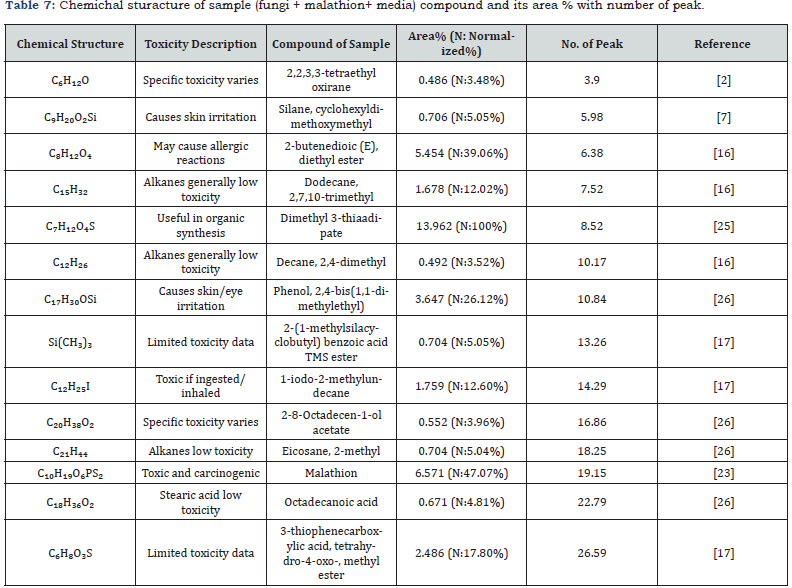
Chemical composition of control and sample
The chemical structures and relative abundances of compounds in the control and sample were analyzed (Tables 6 & 7). The control contained a high concentration of malathion (79.473%), along with other toxic compounds such as phosphorodithioic acid, OO, S-trimethyl ester. In contrast, the sample treated with Alternaria alstroemeriae KRMY EMIC001 showed a significant reduction in malathion concentration (6.571%) and the presence of less toxic degradation products. These findings highlight the potential of Alternaria alstroemeriae Table 6. Chemichal sturacture of controle(malathion + media) compound and its area% with number of peak. KRMY EMIC001 to mitigate environmental pollution by converting toxic pesticides into less harmful compounds [27-40].
Conclusion
Alstromeriae KRMY EMIC001 demonstrates significant potential for malathion bioremediation, achieving >85% degradation under optimized conditions. ITS-based phylogenetic analysis confirmed its identity, while enzymatic and GC-MS data elucidated a phosphatase-driven degradation pathway. This aligns with sustainable strategies for mitigating pesticide pollution, emphasizing the strain’s applicability in contaminated environments
Conflict of Interest Statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Polge J, Robert J, Traon YL (2021) Permissioned blockchain frameworks in the industry: A comparison. ICT Express 7(2): 229-233.
- Tony AM, El Geundi MS, Hussein SM, Abdelwahab MZ (2017) Degradation of malathion in aqueous solutions using advanced oxidation processes and chemical oxidation. Direct Research Journal of Agriculture and Food Science 5(3): 174-185.
- Huang Y, Xiao L, Li F, Xiao M, Lin D, et al. (2018) Microbial degradation of pesticide residues and an emphasis on the degradation of cypermethrin and 3-phenoxy benzoic acid: A review. Molecules 23(9): 2313.
- Raffa CM, Chiampo F (2021) Bioremediation of agricultural soils polluted with pesticides: A review. Bioengineering 8(7): 92.
- Singh B, Kaur J, Singh K (2013) Microbial degradation of an organophosphate pesticide, malathion. Critical Reviews in Microbiology 40(2): 146-154.
- Anger J, Kintz P, Rennes U De, Cedex R (2009) Analytical difficulties in the characterization of pesticides in blood. Annales of Toxic Analysis 21(3): 131-141.
- Van Wyk E, Bouwman H, van der Bank H, Verdoorn GH, Hofmann D (2001) Persistent organochlorine pesticides detected in blood and tissue samples of vultures from different localities in South Africa. Comparative Biochemistry and Physiology Part C: Toxicology & Pharmacology 129(3): 243-264.
- Fürst P, Fürst C, Wilmers K (1994) Human milk as a bioindicator for body burden of PCDDs, PCDFs, organochlorine pesticides, and PCBs. Environmental Health Perspectives 102(Suppl 1): 187-193.
- Johnson LF, Bossler DP, Bossler VON (1959) Calculation of relative permeability from displacement experiments. Petroleum Transactions AIME 216(1): 370-372.
- Thom C, Church MB (1926) The Aspergilli. Williams & Wilkins.
- Johnson LF, Curl EA, Bond JH, Fribourg HA (1960) Methods for studying soil microflora-plant disease relationships. Burgess Publishing Company.
- Metwally AM (1991) New aspects of physiology of glycoamylase production by Aspergillus niger [Doctoral dissertation, Tanta University].
- Raper KB, Fennell DI (1977) The genus Aspergillus. Robert Erieger Publishing Company.
- Gardes M, Bruns TD (1996) Community structure of ectomycorrhizal fungi in a Pinus muricata forest: Above- and below-ground views. Canadian Journal of Botany 74(10): 1572-1583.
- White TJ, Bruns TD, Lee SB, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: MA Innis, DH Gelfand, JJ Sninsky, TJ White (Eds.), PCR protocols: A guide to methods and applications. Academic Press, pp. 315-322.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28(10): 2731-2739.
- Zhou XW, Zhao XH (2015) Susceptibility of nine organophosphorus pesticides in skimmed milk towards inoculated lactic acid bacteria and yogurt starters. Journal of the Science of Food and Agriculture 95(2): 260-266.
- El-Fiki A, Adly M (2020) Morphological, molecular, and organosulphur compounds characterization in irradiated garlic (Allium sativum) by GC-MS and SCoT markers. Journal of Radiation Research and Applied Sciences 13(1): 61-70.
- Geiser DM, Klich MA, Frisvad JC, Peterson SW, Varga J, et al. (2007) The current status of species recognition and identification in Aspergillus. Studies in Mycology 59(1): 1-10.
- Tawfik MM, Ibrahim ME, Mansour MI (2022) Effectiveness of pesticides and their derivatives on soil fungal biota and role of these fungi in bioremediation of pesticide residues. Alfarama Journal of Basic & Applied Sciences 3(2): 239-252.
- Kulshrestha G, Kumari A (2011) Fungal degradation of chlorpyrifos by Acremonium sp. strain (GFRC-1) isolated from a laboratory-enriched red agricultural soil. Biology and Fertility of Soils 47(2): 219-225.
- Derbalah A, Ismail A (2013) Remediation technologies of diazinon and malathion residues in aquatic system. Environment Protection Engineering 39(3): 135-147.
- Gan J, Koskinen WC (1998) Pesticide fate and behavior in soil at elevated concentrations. In: PC Kearney (Ed.), Pesticide remediation in soils and water. John Wiley & Sons, pp. 59-84.
- National Center for Biotechnology Information (2025) PubChem Compound Summary for CID 13531453, 2-Chlorobenzenesulfonic acid.
- Rønhede S, Jensen B, Rosendahl S, Kragelund BB, Juhler RK, et al. (2005). Hydroxylation of the herbicide isoproturon by fungi isolated from agricultural soil. Applied and Environmental Microbiology 71(12): 7927-7932.
- S. Pharmacopeial Convention (2020) Isomalathion Safety Data Sheet (SDS), Version 04. USP.
- Abd Naser HS, Sani I (2008) Organochlorine pesticide residues in the major rivers of Southern Thailand. Malaysian Journal of Analytical Sciences 12(2): 280-284.
- Agency for Toxic Substances and Disease Registry (2017) Toxicological profile for malathion. U.S. Department of Health and Human Services.
- American Public Health Association (APHA) (2017) Standard methods for the examination of water and wastewater (23rd edn).
- Australian Industrial Chemicals Introduction Scheme (AICIS) (n.d.). Octadecanoic acid.
- CAS Common Chemistry. (n.d.). 1,4-Di(ethyl-1,1,2,2,2-d5) 2 [(dimethoxyphosphinothioyl)thio]butanedioate.
- Deiz MC (2010) Biological aspects involved in degradation of organic pollutants. Journal of Soil Science and Plant Nutrition 10(3): 244-267.
- de Souza JC, de Carvalho Couto C, Mamede AMGN, Valderrama P, et al. (2024) Using volatile compounds for the identification of coffee adulterants: Marker compounds and non-targeted analysis. European Food Research and Technology 250: 2639-2649.
- El-Abd SBH, Abu-Shady H, Elshebiny HAFM, Ebrahim MAA, Sayed HA (2021) Malathion biodegradation by Lactobacillus casei (NRRL1922) and Lactobacillus acidophilus (NRRL 23431) in fermented skimmed milk. Journal of Pure and Applied Microbiology 15(3): 1617-1624.
- Frinton Laboratories Inc (1986) Catalog Number 11. Vineland, NJ.
- Gianfreda L, Rao MA (2004) Potential of extracellular enzymes in remediation of polluted soils: A review. Enzyme and Microbial Technology 35(4): 339-354.
- Hamilton D, Crossley S (2004) Pesticide residue in food and drinking water: Human exposure and risks. John Wiley & Sons.
- Majewski M, Snieckus V (1982) Ortho-Metalation of O-aryl N,N-dialkylcarbamates. Directed ortho lithiation of O-aryl N-monoalkylcarbamates. Journal of the American Chemical Society 104(23): 6122-6128.
- Moorthy NM, Umetsu N, Toia RF, Talcott RE, Roy FT (1979) Toxicity of O,O,S-trimethyl and triethyl phosphorothioate to the rat. Journal of Agricultural and Food Chemistry 27(2): 463-466.
- Oliveira CT, Alves EA, Todero I, Kuhn RC, de Oliveira D, et al. (2019) Production of cutinase by solid-state fermentation and its use as adjuvant in bioherbicide formulation. Bioprocess and Biosystems Engineering 42(5): 829-838.






























