Abstract
The gut microbiome encompasses not only bacteria but also viruses (the virome) and fungi (the mycobiome), which play crucial roles in gut homeostasis and host health. However, research has predominantly focused on the bacterial component, with the virome and mycobiome remaining comparatively underexplored due to their lower abundance and the challenges associated with their detection. Emerging evidence indicates that these components significantly contribute to mucosal barrier integrity, metabolic processes, and immune modulation and that disturbances in the gut virome or mycobiome (dysbiosis) are associated with various diseases, including inflammatory bowel disease, metabolic disorders, and autoimmune conditions. In this review, we examine the composition and functions of the gut virome and mycobiome in both healthy and diseased contexts, highlighting their interactions with each other and with bacterial communities. We discuss how bacteriophages shape bacterial populations and how fungi such as Candida influence metabolism and immunity. We also address recent advances in viral and fungal metagenomics that have begun to illuminate the extensive “dark matter” of the gut virome and the diversity of the mycobiome. Finally, this review identifies critical gaps in current knowledge and outlines future directions for integrating virome and mycobiome insights into microbiome research and therapeutic strategies.
Keywords:Gut Microbiome, Bacteria, Gut virome, Clinical Significance, Disease, dysbiosis, mycobiome, Bacteroides
Introduction
Over the past decade, our understanding of the human gastrointestinal (GI) microbiome has expanded significantly, with new insights into the trillions of microorganisms residing in the gut [1]. These microbes, including bacteria, viruses, and fungi, interact closely with the host environment and are essential to human health, participating in digestion, breakdown of polysaccharides, vitamin synthesis, and protection against pathogens [2,3]. They also play a significant role in immune regulation and communication with host cells [4,5]. Disruption of the gut microbiome (dysbiosis) is now recognized as a key factor in many diseases, not only in classic infections but also in obesity, diabetes, liver disorders, and cancer [2,4,6]. Consequently, the gut microbiota has emerged as a promising target for new diagnostic and therapeutic approaches. While bacteria are the most abundant and well-studied constituents of the microbiome, the gut also harbors important populations of viruses (the virome) and fungi (the mycobiome), both of which contribute to mucosal barrier function and gut homeostasis [7-9]. Research has focused predominantly on the bacterial fraction, largely because viruses and fungi are less abundant and more complex to identify using standard laboratory methods [10-12]. However, advances in sequencing and bioinformatics are beginning to shed light on the roles of these previously overlooked communities. This review examines the current understanding of the gut virome and mycobiome in human health and disease, with an emphasis on their roles in gut homeostasis, immune modulation, and disease mechanisms. This study precisely reviewed publications addressing the composition, function, or clinical implications of the intestinal virome or mycobiome, prioritizing recent and mechanistic work.. This synthesis highlights key knowledge gaps, advances in methodology, and future directions for research and clinical application.
Methodology
In this review, a narrative search of PubMed, Scopus, and Google Scholar using the terms “gut virome”, “gut mycobiome”, “viral microbiome”, “fungal microbiome”, and “microbiome dysbiosis in health and disease” was performed. Additionally, other information with respect to study aims was also collected. No date limits were imposed to capture both foundational and contemporary work; however, preference was given to studies published within the past decade and those offering mechanistic insights or clinically applicable findings, and pros and cons were discussed accordingly.
Gut virome: composition, function, and clinical significance
Definition and components of the Gut Virome
The gut virome often described as the “dark matter” of the gut
microbiome [13], is a genetically diverse and complex microbial
community. Various body sites, including the blood, nasal passages,
skin, conjunctiva, oral cavity, vagina, and gastrointestinal (GI)
tract, each harbor their own distinct populations of viruses [14]
The virome comprises eukaryotic viruses that primarily infect human
cells, prokaryotic viruses known as bacteriophages (phages),
and archaeal viruses [15,16]. In the human gut, bacteriophages
are by far the most abundant, outnumbering eukaryotic viruses,
and are the predominant viral group present [17,18]. Fecal samples
contain between 10⁹ and 10¹⁰ viral particles per gram, reflecting
the remarkable density and persistence of these phages
within the gut environment [13]. Although eukaryotic viruses are
less prevalent, they have been the focus of significant research
due to their role in intestinal diseases. Additionally, the human
genome contains rare endogenous retroviruses, which can influence
disease processes by altering gene expression and immune
regulation [19].
Role in Gut Homeostasis The human gut virome plays a vital
role in both human health and ecological biology [20]. Colonization
of the gut virome begins at birth. It gradually stabilizes into
adulthood, with each individual developing a unique virome profile
influenced by factors such as age, antibiotic use, and underlying
health conditions [21]. Even the mode of delivery can shape
the initial virome composition: newborns delivered vaginally
have a more diverse gut virome compared to those delivered by
cesarean section, with Caudoviricetes, Microviridae, and Anellovi
ridae among the most abundant viruses detected [22]. Moreover,
specific changes in the virome have been linked to various disease
states, including inflammatory bowel disease, diabetes, hypertension,
AIDS, colorectal cancer, and acute malnutrition [17].
These associations reflect the complex, multifaceted interactions
between the virome, particularly bacteriophages, and the gut bacteriome,
which together contribute to microbial colonization, ecological
balance, and host health through dynamic host-pathogen
interactions [23].
Gut virome in Disease
Although research into the gut virome remains in its early stages, emerging evidence reveals that disruptions in viral community composition and reduced diversity can impair intestinal barrier function. A notable pathogenic mechanism involves viruses crossing the gut epithelium, especially in conditions with increased permeability, which may trigger immune responses and contribute to autoimmune, infectious, and inflammatory diseases such as inflammatory bowel disease (IBD) [14,24]. The pathogenesis of IBD is multifactorial, involving genetic susceptibility, immune activation, and environmental influences—including the gut microbiota [25,26]. Animal studies further suggest that certain eukaryotic viruses can interact with host genetic risk factors to modify intestinal disease risk [27]. In healthy individuals, the dominant gut phages are double-stranded DNA (dsDNA) Caudovirales and single-stranded DNA (ssDNA) Microviridae [28]. When exposed to environmental stressors like nitric oxide or antibiotics, phages may switch to the lytic cycle, causing bacterial lysis and spreading infectious virions within the gut [29]). Increased Caudoviricetes abundance has been observed in both ulcerative colitis and Crohn’s disease, while overall phage diversity is lower in Crohn’s [30,31]. Distinct virome changes are also noted in metabolic and inflammatory diseases. For example, shifts in the gut phagosome have been reported in obesity, type 1 and type 2 diabetes [32,33] though whether these are causal or simply reflect broader dysbiosis is unclear. Notably, the human gut microbiome accounts for a significant portion of resting energy expenditure, and disturbances in its composition may contribute to metabolic dysfunction [34]. In pregnant women with early-onset diabetes, increased levels of tobamoviruses and picobirnaviruses have been detected; these changes may serve as biomarkers of immune alterations, though their precise roles and host targets remain uncertain [21]. COVID-19, caused by SARS-CoV-2, further illustrates the clinical relevance of the gut virome. The virus infects the gastrointestinal tract via the ACE2 receptor, and patients even those with mild disease show reduced virome diversity and an increase in inflammation-associated viral genes [21,35]. While short-term changes are well described, the long-term impact on gut health requires further study. Ultimately, the virome, along with the bacteriome and mycobiome, forms a tightly interconnected network in the gut. Disruptions in one component can reshape the entire microbial ecosystem and influence host immunity and disease risk. For example, changes in phage populations can affect bacterial evolution, while virome alterations have been implicated in autoimmunity, such as rheumatoid arthritis, where distinct shifts in specific bacteriophages have been observed in at-risk individuals [7,8].
Challenges and advances in virome research
Bioinformatics challenges in viral metagenomics stem primarily from the phenomenon known as “viral dark matter,” which refers to the 40–90% of gut viruses in a sample that remains unidentified and unexplored [36]. This uncharacterized genetic material holds substantial potential for discoveries; for instance, analysis of this dark matter led to the identification of crAssphage, now known as one of the most common bacteriophages in the human gut. Addressing this gap requires the development of improved computational tools. Methods such as hidden Markov models, which detect distant similarities in viral proteins, are being used to classify unknown viruses. Advances in machine learning and artificial intelligence, including neural networks, further improve classification accuracy. Experimentally, culturing more viruses in the laboratory will generate valuable reference data and help reduce the number of unclassified sequences [37]. As a result, advancing our understanding of the gut virome is crucial for uncovering its role in human health. Research on the gut virome remains limited, mainly due to incomplete reference libraries and technological hurdles [38]. Many intestinal viruses remain unclassified, but ongoing improvements in metagenomic technologies, including the combination of short-read and long-read sequencing, are expected to overcome these barriers [39]. Enhanced, cost-effective, and sensitive sequencing methods now allow for more comprehensive characterization of the human virome, encompassing double-stranded DNA (dsDNA), single-stranded DNA (ssDNA), and RNA virus-like particles [14]. This progress supports the exploration of diverse gut bacteriophage populations and aids in the discovery of novel viral genomes [40]. To support these efforts, several human gut virome databases have been established, including the Gut Virome Database (GVD), the Cenote Human Virome Database (CHVD), the Metagenomic Gut Virus Database (MGV), and the Gut Phage Database (GPD) [41]. Most well-known databases encompass the Gut Virome Database (GVD), Cenote Human Virome Database (CHVD), Metagenomic Gut Virus Database (MGV), and Gut Phage Database (GPD). These resources have significantly expanded our understanding of gut viral diversity, providing accurate and informative datasets that serve as guidelines for future studies [42].
The Gut Mycobiome: Diversity, Function, and Pathogenic Potential
The human gut microbiome the collection of microorganisms in the gastrointestinal tract - is increasingly recognized as a key player in the pathogenesis of various diseases, including inflammatory bowel disease (IBD) [43], type 2 diabetes [44], hypertension [45], and colorectal cancer [46]. Both bacteria and fungi contribute to host metabolism, with research highlighting asso ciations between microbial populations and metabolic outcomes in conditions like diabetes [47] and cholesterol metabolism [48]. Lifestyle factors such as diet and physical activity substantially influence the gut microbial community, as demonstrated in animal models and human cohort studies [49,50].
The gut microbiota behaves in a metabolically active manner, operating symbiotically with the gut mucosa to contribute to metabolic, immunological, and protective functions [51]. Current research no longer focuses solely on enumerating the abundance and diversity of gut microbes but rather on how they contribute to overall health. Most nutrients for the gut microbiota are derived from dietary carbohydrates, fermented by colonic bacteria such as Bacteroides, Roseburia, Bifidobacterium, Fecalibacterium, and Enterobacteria to produce short-chain fatty acids (SCFAs) [52,53]. The microbiota also synthesizes essential vitamins (K and B groups) and conjugated linoleic acid (CLA), which have anti- inflammatory, antiatherogenic, and immunomodulatory effects [54]. Additionally, specific bacteria, including Bacteroides intestinalis, Bacteroides fragilis, and E. coli, transform primary bile acids into secondary bile acids, such as deoxycholic and lithocholic acid [55]. The gut microbiome’s influence on drug metabolism has long been recognized; for example, the microbial metabolite p-cresol inhibits acetaminophen metabolism, and microbial activities can increase toxicity from drugs like irinotecan, resulting in side effects such as diarrhea and inflammation [56-58].
Maintaining the balance of the gut microbiota requires the mucosal immune system to tolerate commensals while preventing the overgrowth of pathogens. This is achieved through a dual-layer mucus barrier produced by goblet cells, which shields the epithelium from microbes. The inner mucus layer is dense and typically free from microorganisms, while the outer layer provides nutrients and stability to the mucin polymers, thereby supporting barrier integrity [59-61]. Both innate and adaptive immune components, including gut-associated lymphoid tissues, effector T cells, and innate lymphoid cells, work in conjunction with the microbiota to modulate [62] immune responses [63].
Currently, there is a convincing body of evidence that supports the role of the gut microbiota in maintaining the structure and function of the gastrointestinal tract. Bacteroides thetaiotaomicron is reported to induce expression of the small proline-rich protein 2A (sprr2A), which is required to maintain desmosomes at the epithelial villus [64]. External and host factors can induce dysbiosis in the gut microbiome, impairing host wellness and potentially leading to harmful microbial-derived products or metabolites, causing various diseases in local, systemic, or remote organs, and requiring microbiome-based therapy [65].
Clostridium difficile, a Gram-positive anaerobe and member of the Firmicutes, produces toxins A and B that damage colonic epithelial barrier integrity, causing an inflammatory response and cell death [66,67]. C. difficile infection (CDI)-associated symptoms include diarrhea, pseudomembranous colitis, sepsis, and death in severe cases [68].
Another example of gut microbiome-associated disease is IBD. IBD is a group of multifactorial, idiopathic, persistent, and recurring gastrointestinal inflammations. Two common forms of IBD are celiac Disease (CD) and ulcerative colitis (UC) [69]. CD and UC inflammations, causing relapsing diarrhea, fever, and abdominal pain, are increasing globally, affecting 1.4 million and 2.2 million individuals in America and Europe, respectively [70]. IBD is a complex disease influenced by host and environmental factors, with gut microbiota and host factors potentially intertwining to contribute to its development [65].
The gut microbiome, a key factor in host metabolism regulation, has been linked to obesity, a global health hazard affecting over 600 million people. Obesity is associated with high energy intake, metabolic syndrome, obesity-related disorders, low-grade inflammation, and premature mortality [71].
Composition of the Gut Mycobiome Common Fungal Species
Animal models reveal the role of intestinal fungi, particularly Candida, in energy harvest and host metabolism. Candida albicans and Candida parapsilosis have been studied extensively, showing contrasting effects on metabolic hormones and obesity development, highlighting the need for further research [62,72,73].
The human gut mycobiome is typically dominated by commensal yeasts such as Saccharomyces cerevisiae, Malassezia restricta, and Candida albicans, which coexist with bacteria to support digestion, modulate immune function, and prevent the overgrowth of pathogens. Analysis of stool samples from healthy individuals in the Human Microbiome Project showed that fungal diversity is lower than bacterial diversity, with yeast, particularly Saccharomyces, Malassezia, and Candida, being the most prevalent genera. The persistence of particular fungal species across multiple individuals suggests the existence of a core gut mycobiome [62,74].
Mycobiome-Host Interactions Cross-talk between fungi, bacteria, and immune system Role in digestion and metabolism
Bacterial-fungal interactions in the host are shaped by immune responses, with certain microbial colonizers inducing significant immunological changes that influence the balance of other microbes [75]. For example, Bacteroides thetaiotaomicron activates innate immune genes in mice, which suppresses colonization by Candida albicans. Overall, studies indicate that both bacterial and fungal species can have important effects on the host immune system [76].
Recognition of fungi by the immune system
The host’s immune response to fungi relies on immune cells detecting molecular patterns through pattern recognition receptors (PRRs), including Toll-like receptors, C-type lectin receptors, and NOD-like receptors. Phagocytosed fungi can activate NOD-like receptors (NLRs), triggering inflammation [77,78]. Mutations in the Dectin-1 gene increase susceptibility to Candida tropicalis invasion and worsen colitis in both mice and humans, while mutations in Dectin-2 are linked to higher rates of Candida glabrata infection [79]. deficiencies in TLR4 and TLR2 signaling impact systemic candidiasis outcomes. Specifically, TLR4 deficiency leads to an increased C. albicans burden in the kidneys, while blocking TLR2 reduces inflammatory cytokine production, including TNF-α and IL-1β [80,81]. Other receptors, including MelLec, pentraxins, MBL, and MDA5, are also involved in fungal recognition, such as binding to Aspergillus fumigatus conidia, recognizing galactomannan and mannan, and mediating immune responses to systemic Candida albicans infection [82,83].
Immune system-mycobiota interaction
Research indicates that fungi play a crucial role in immune system maturation, as their presence enhances circulating granulocyte levels in mice residing in natural environments [84]. This builds on earlier work indicating that mice colonized with a “wildmouse microbiota” exhibit immune responses to immunotherapy that more closely resemble those of humans [85]. Fungi, and the mycobiome in general, are vital for immune system development and homeostasis but can also play a role in inflammatory and pathogenic conditions. For example, Candida and Malassezia, especially C. albicans, can exacerbate gut inflammation [86]. Fungal dysbiosis and increased colonization by C. albicans have been linked to IBD in humans, underscoring the importance of understanding the mycobiome’s role in immune system development [87].
Gut Mycobiome in Disease Crohn’s disease
Crohn’s disease (CD) is a chronic, severe inflammatory condition that most often affects the terminal ileum and can cause irreversible damage throughout the gastrointestinal tract, leading to significant reductions in quality of life [88]. Its pathogenesis involves a combination of genetic, epigenetic, immunological, and microbiological factors, as well as environmental triggers like smoking, diet, stress, and infections. Overgrowth of microbes, such as Mycobacterium avium and adherent-invasive Escherichia coli (AIEC), may also play a role [89]. Current evidence suggests that dysbiosis—an alteration in the gut microbiome may be either a cause or a consequence of CD, with affected patients displaying reduced microbial community complexity compared to healthy individuals [90]. Faecalibacterium prausnitzii, a Firmicutes bacterium with proposed anti-inflammatory effects, has been found at lower levels in CD patients and may be linked to a higher risk of disease recurrence [91,92].
Irritable bowel syndrome (IBS)
Irritable bowel syndrome (IBS) is a common functional gastrointestinal disorder that, although rarely life-threatening, imposes a substantial socioeconomic burden and significantly reduces quality of life [93]. IBS is diagnosed based on persistent and recurrent symptoms, and while treatment focuses on symptom relief, atypical presentations can complicate management over time [94]. The pathogenesis of IBS is increasingly linked to micro- dysbiosis alterations in the gut microbial community with more than 70% of patients showing reduced diversity and distinct microbial profiles compared to healthy controls. Notably, IBS is often associated with increased levels of facultative anaerobes, such as Streptococcus species, and decreased levels of Lactobacilli and Bifidobacteria. Many studies also report an elevated fecal Firmicutes/ Bacteroidetes ratio among IBS patients [95].
Colorectal cancer (CRC)
Colorectal cancer (CRC) ranks as the fourth leading cause of cancer mortality worldwide. It arises from a combination of genetic and environmental influences, with the gut microbiota playing a key role in promoting a tumor-friendly environment. Experimental models, such as those using APC mutations, demonstrate the role of the microbiome in colorectal carcinogenesis [96]. There is a complex, bidirectional relationship between the gut microbiome and CRC: gut fungi interact with the host immune system, modulating immunity and potentially contributing to conditions such as IBD, asthma, and cancer [97]. The bacterial microbiome also influences host metabolism by shaping immune cell populations and inflammatory tone, with intestinal fungi potentially affecting host energy balance through their immunomodulatory properties [98].
Challenges and Future Directions in Mycobiome Research Fungal metagenomics and biomarker discovery
While advances in metagenomics and experimental techniques have expanded our understanding of the mycobiome, significant technical and conceptual challenges remain. Low fungal biomass and lack of control over data sources contribute to variability in study results, particularly when investigating the mycobiome’s role in cancer [99]. Many studies do not provide detailed protocols for sample contamination control, highlighting the need for more rigorous sample collection and larger, well-defined cohorts. Current fungal databases are limited by inconsistent species nomenclature and insufficient data; therefore, harmonization of bioinformatics analysis tools is necessary for improved data comparability [100]. The reproducibility and accuracy of pan-cancer mycobiome analyses have not been fully validated, making future research with improved coordination, robust data sharing, and standardized denoising procedures essential for their validation. Study design, specimen source, population characteristics, cancer type, pathological subtype, and environmental factors can all affect study outcomes and interpretations [62]. High inter-individual variability is common in mycobiome studies and may contribute to immune-mediated disorders driven by rare fungal species. However, this variability could also offer insights for predicting individualized disease changes [101]. Future cancer mycobiome research should prioritize causality, collaborative work, and mechanistic understanding. While correlation studies are valuable, experimental validation through controlled experiments and extensive model validation is essential for establishing causal links between fungi and cancer [102]. Animal models and other experimental systems can help simulate cancer environments, validate microbiome profiles, and assess the effects of fungi on tumor development. New technologies also allow the spatial distribution of fungal communities within tumor tissues to be studied [103]. Inter-kingdom interactions between fungi and bacteria have been documented in cancer, suggesting that communities of multiple microbial species, rather than individual ones, may collectively drive ecological dysbiosis [104]. Most research to date relies on metagenomics, but integrating meta-transcriptomics and meta-proteomics in a multi-omics approach could lead to the identification of more reliable biomarkers [105]. Clinical studies indicate that combining chemotherapy or immunotherapy with microbiome modulation may enhance cancer management. However, mycobiome research has yet to be translated into clinical therapeutic interventions for humans [106].
Gut Virome-Mycobiome Interactions: The Unexplored Cross-Talk
Influence on bacterial microbiome
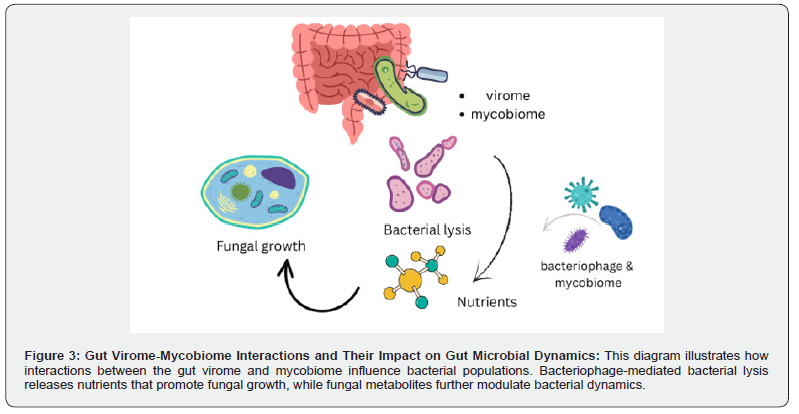
The gut virome, particularly bacteriophages, interacts with gut bacteria, shaping the gut microbiota [107] and altering bacterial gene expression, impacting the host’s immune response [108]. This interaction also leads to microbial imbalances, such as in IBD [109]. Although the gut mycobiome is significantly smaller than the bacterial population, it has a substantial impact on bacterial colonies by producing metabolites that inhibit bacterial growth and by competing with bacteria for nutrients and space [110]. The virome can also transfer antimicrobial resistance genes to bacterial pathogens, enabling them to develop drug resistance [111]. Interactions between the virome and mycobiome can be either synergistic or antagonistic. For example, phage-induced bacterial lysis, supported by fungal metabolites, releases nutrients that may promote fungal growth, illustrating the complex dynamics that shape gut ecology [112]. These multi-kingdom interactions among viruses, fungi, and bacteria are essential for maintaining the structure and function of the gut microbiome. Disruptions in these interactions can lead to microbial imbalances, which in turn affect host health and increase disease susceptibility [111].
Implications for gut inflammation and systemic diseases
Imbalance in the gut microbiota including disruptions in the virome and mycobiome-can drive systemic inflammation and is implicated in diseases such as rheumatoid arthritis and systemic lupus erythematosus [113]. Dysbiosis of the virome can trigger immune responses that promote gut inflammation and contribute to IBD, characterized by a loss of microbial diversity and the accumulation of specific bacterial groups, such as the Ruminococcus gnavus clade and E. coli [109,114,115]. Alterations in the mycobiome, particularly the overgrowth of Candida, can further escalate gut inflammation and disease by stimulating the host immune system [116]. The virome and mycobiome also regulate the production of microbial-derived metabolites, which can affect host metabolism. Their dysbiosis can lead to chronic low-grade inflammation, contributing to metabolic syndromes such as obesity and type 2 diabetes mellitus [117]. Additionally, the gut microbiota influences lung diseases-for example, lipopolysaccharide stimulation in atopic asthma and gut injury or inhibited epithelial growth in pneumonia. Recent findings suggest that altering the gut microbiota can lead to metabolites that promote inflammation in distant organs such as the lungs and bone marrow. The mycobiome not only affects local gut inflammation but can also contribute to systemic inflammation by releasing metabolites that impact distant tissues and organs.
Potential therapeutic targets
Emerging therapeutic strategies now focus on developing microbiome-modulating drugs, such as those targeting bacteriophage- host interactions, as new treatments for chronic gastrointestinal diseases [108]. Diet remains a central factor in regulating the gut microbiome; for example, a fiber-rich diet in patients with type 2 diabetes mellitus (T2DM) has been shown to improve insulin sensitivity by promoting beneficial gut fermenters and enhancing glucose metabolism. Similarly, supplementation with flaxseed mucilage in obese women resulted in changes to the gut microbiome and improvements in glycemic control [118]. Bacteriophage therapy represents another promising approach, demonstrating efficacy in treating antibiotic-resistant bacterial infections in clinical settings [119]. Modulating the gut mycobiome with antifungal agents has also been shown to reduce inflammation in conditions such as Crohn’s disease and ulcerative colitis [109]. Targeting the virome, particularly bacteriophages, to restore microbial ecosystem balance has been linked to symptom improvement in inflammatory bowel disease [107]. Microbiota transplantation is another strategy, with evidence showing that transferring gut microbiota from healthy donors can alleviate both gastrointestinal and behavioral symptoms in patients with autism spectrum disorder [118]. Combining virome and mycobiome modulation therapies offers a synergistic approach to reestablishing microbial balance and treating inflammatory conditions [120].
Current and future therapeutic approaches.
Virome-Based Therapies
The “human virome” encompasses all viruses present on or within the human body, including those responsible for acute, persistent, or latent infections, as well as endogenous retroviruses integrated into the genome [121]. While microbiome research has historically centered on bacterial communities due to well-established techniques for bacterial study, the archaeal, eukarya, and viral components have received less attention [122]. Recent advancements in high-throughput sequencing and bioinformatics have expanded and standardized virome research, bringing new insight into the gut virome’s role in health and disease [123,124].
As the relationship between gut dysbiosis and disease becomes clearer, targeted manipulation of the gut virome is emerging as a therapeutic focus. Approaches such as prebiotics, probiotics, synbiotics, fecal microbiota transplantation (FMT), phage therapy, and fecal virome transplantation (FVT) are increasingly explored. Phage therapy stands out for its ability to specifically target and eliminate pathogenic bacteria, making it especially valuable for infections caused by invasive or drug-resistant organisms [123]. Phage therapy and FVT are now being studied as promising alternatives for treating Clostridioides difficile infections, particularly those involving resistant strains. Given the risks associated with broad-spectrum antibiotic therapy, complementary strategies such as FMT, FVT, and phage-based interventions are being adopted to treat, prevent, and reduce the recurrence of CDI [125].
As virome research and bioinformatics advance, improved databases and analytical tools are enabling the achievement of strain-level resolution, thereby facilitating more accurate identification of dysbiosis-associated viral communities [124]. Thus, virome-based therapies represent a rapidly growing frontier in microbiome interventions, with significant potential for treating a broad range of diseases
Phage therapy for gut dysbiosis
A balanced gut microbiome is essential for maintaining intestinal barrier integrity and preventing pathogenic invasion [126]. Dysbiosis-disruption of microbial composition and function-may result from various factors, including diet, toxins, drugs, and pathogens [127]. This imbalance alters microbial populations and metabolism, disrupts the gut barrier [128] and can lead to various inflammatory and systemic diseases [129]. Phage therapy, which utilizes bacteriophages to target specific harmful bacteria, has emerged as a promising strategy for restoring microbial balance [129]. Phages are highly specific to their bacterial hosts, replicate using bacterial machinery, and do not persist outside their target bacteria [130]. Therapeutic approaches include conventional and engineered phages, as well as phage-derived enzymes [131]. The gut virome predominantly includes ssDNA phages (Microviridae) and dsDNA phages [132].
Metagenomic studies have identified crAssphage, a dsDNA phage, as highly prevalent in the human gut, present in approximately half of individuals, and constituting up to 90% of viral DNA in stool [133]. Healthy adults tend to share a core set of bacteriophages, forming the healthy gut phagosome (HGP), which is notably reduced in diseases such as ulcerative colitis and Crohn’s disease (by up to 42–54%) [134].
Phage therapy aims to modulate the phageome to beneficially influence the bacteriome. This involves selecting or engineering phages for specific bacterial targets, preparing effective phage formulations, and establishing appropriate dosing strategies [135]. While the introduction of antibiotics historically overshadowed phage therapy, the rise of antibiotic resistance has renewed interest in these targeted interventions [136]. Phages offer strain specificity, which reduces the risk of complications like antibiotic- associated diarrhea and Clostridium difficile infection [137]. Their ability to replicate within the host also enables sustained therapeutic activity at the site of infection [138]. Recent research highlights the potential of phage therapy. For instance, a fivephage cocktail targeting Klebsiella pneumoniae in IBD models reduced inflammation and disease severity [139]. Furthermore, the selection of bacteriophages from families such as Leviviridae, Microviridae, and Caudovirales (Myoviridae, Siphoviridae, and Podoviridae) underlines their broad applicability in clinical contexts [140]. Clinical applications use phages from families such as Leviviridae, Microviridae, and Caudovirales. Phages can also work synergistically with antibiotics, disrupting bacterial biofilms and improving antibiotic penetration 140,141,142. Safety and efficacy have been demonstrated in human trials. Oral administration of T4-like phage cocktails in children with acute bacterial diarrhea confirmed the safety of phages [143]. The PHAGE study in healthy adults found a four-strain phage cocktail to be safe and well-tolerated [144]. Another trial demonstrated that phage supplementation selectively reduced fecal E. coli without affecting overall microbiota diversity and was associated with favorable shifts, including increased butyrate-producing Eubacterium and reduced Clostridium perfringens [145]. Animal studies also confirm the efficacy of phages in reducing pathogenic bacteria in vivo [146]. Phage therapy can selectively eliminate pathogenic species using lytic phages or potentially induce temperate phages in commensals; however, the latter approach is limited by an incomplete understanding of phage induction mechanisms [147]. Targeted phage therapies have shown success in models of IBD; for example, a seven-lytic-phage cocktail targeting adherent invasive Escherichia coli (AIEC) reduced inflammation and preserved non-target commensal strains [148]. Collectively, these studies demonstrate the promise of phage therapy as a precise and effective tool for modulating the gut microbiota and addressing dysbiosis. Further research is needed to optimize dosing and better understand phage-immune system interactions, but phage therapy is an emerging alternative to antibiotics for gut health.
Antiviral Interventions for Gut Health
Recent research suggests that antiviral treatments can decrease viral loads and have a positive impact on the gut microbiota. Direct-acting antiviral agents (DAAs), for example, are effective in eradicating viruses and have been linked to a partial restoration of α-diversity and a reduction in potentially pathogenic species; however, these benefits are less evident in patients with cirrhosis [149]. In inflammatory bowel disease, particularly Crohn’s disease, an increase in Caudovirales has been linked to reduced bacterial diversity, suggesting that virome alterations may contribute to intestinal inflammation and dysbiosis [150]. During viral infections, epithelial and innate immune cells recognize pathogens via pattern recognition receptors, activating inflammatory cascades that generate type I and III interferons, IL-6, IL-1β, IL-18, and TNF-α—key mediators in antiviral defense and leukocyte recruitment [151]. Beyond antiviral drugs, immune modulation with probiotics offers additional benefits. Probiotic supplementation can strengthen the gut’s antiviral defenses by influencing microbial composition and stimulating interferon production, potentially supporting longer-term resistance to infections like COVID-19 [152]. In HIV, INSTI-based antiretroviral regimens promote better recovery of gut microbiota diversity compared to NNRTI-based therapies, underscoring the relationship between antiviral treatment and microbiome restoration [153]. Vitamin A supplementation has also been shown to inhibit murine norovirus replication and increase the abundance of Lactobacillus, further enhancing antiviral protection [154]. Collectively, these findings highlight the multifaceted impact of antiviral interventions not only in reducing viral pathogens directly but also in supporting a healthier and more resilient gut microbiota.
Mycobiome-Targeted Interventions
The healthy gut mycobiome, primarily composed of commensal fungi such as Saccharomyces cerevisiae, Malassezia restricta, and Candida albicans, is essential for intestinal homeostasis— supporting digestion, modulating immunity, and preventing pathogenic overgrowth [101]. Disruption of the mycobiome, or fungal dysbiosis, has been increasingly linked to a range of diseases, including gastrointestinal, metabolic, neurological, and cardiovascular disorders [155]. Recognizing its importance, the mycobiome is now viewed as a promising target for diagnostics and therapy in precision medicine. Strategies to restore mycobiome balance include the use of probiotic fungi, antifungal medications, dietary modifications, and fecal microbiota transplantation (FMT) all of which have shown potential to re-establish a healthy gut environment [155,156]. Ongoing research into these interventions may yield new treatments for mycobiome-associated diseases.
Antifungal Strategies and Probiotics
Fungal infections cause over 1.5 million deaths globally each year, particularly in individuals with weakened immune systems [157]. The increasing incidence of these infections, combined with rising resistance and side effects from conventional antifungal drugs, highlights the urgent need for safer, broad-spectrum alternatives [158,159]. Probiotics live microorganisms that confer health benefits when administered in adequate amounts have emerged as promising antifungal agents by boosting host immunity and preventing pathogen colonization [159]. Research has demonstrated that specific probiotic strains can inhibit the growth of pathogenic fungi. For example, certain probiotics are effective in preventing and treating dermatophyte infections [160]. Lactobacillus reuteri R2, in particular, showed potent antifungal activity: its freeze-dried supernatant, at concentrations above 1%, completely inhibited the growth and germination of Trichophyton tonsurans [161]. Another study found that combining probiotics with seaweed extract (Ascophyllum nodosum) produced significant in vitro antifungal effects against Trichophyton mentagrophytes and Candida albicans [162].
Additional investigations have highlighted the efficacy of various probiotic bacteria, including Streptococcus salivarius K12, Lactobacillus rhamnosus GR-1, Lactobacillus reuteri RC-14, and clinical Lactobacillus isolates, against Candida species [163]. Clinical studies support these findings: In preterm infants, supplementation with Lactobacillus species has been shown to reduce gastrointestinal Candida colonization, decrease late-onset sepsis, and improve neurological outcomes [164]. Similarly, a randomized, double-blind trial found that probiotic supplementation reduced rates of fungal colonization and invasive fungal sepsis, promoted earlier initiation of full enteral feeds, and shortened hospital stays compared with the placebo [165]. These results support the use of probiotics as an antifungal strategy either alone or in combination with traditional agents—to control fungal infections and help maintain a balanced mycobiome.
Holistic Microbiome Modulation Fecal Microbiota Transplantation (FMT), Including Virome and Fungi
Fecal microbiota transplantation (FMT) involves transferring the microbiota from the stool of a healthy donor to a patient with gut dysbiosis. In addition to bacteria, the human gut contains a diverse virome and mycobiome-collections of viruses and fungi, respectively [166]. While FMT is primarily used to treat Clostridioides difficile infection (CDI), recent research highlights the critical role of the virome and mycobiome in its efficacy.
CDI patients exhibit an enrichment of bacteriophage Caudovirales, characterized by reduced viral diversity compared to healthy individuals. Successful FMT reduces the abundance of Caudovirales, and the presence of donor-derived Caudovirales in recipients correlates with clinical cure [167]. Donor-derived bacteriophages can persist in recipients for up to a year, with long-term outcomes influenced by the compatibility between the donor and recipient [168]. Refined approaches such as fecal viral transfer (FVT) and fecal filtrate transplantation (FFT) aim to transfer virome components without bacteria, potentially reducing infection risks. However, the persistence of eukaryotic viruses and prophage-encoded virulence factors requires caution [169]. Animal studies suggest that FVT can help restore gut homeostasis after antibiotics, with recipient mice regaining pre-antibiotic bacteriome profiles more effectively when given viable virome transplants [170]. Fungal constituents of FMT are also gaining attention for their immunomodulatory and therapeutic roles. In patients with ulcerative colitis (UC), FMT-induced remission was associated with an increase in beneficial fungi (e.g., Kazachstania naganishii, Schizosaccharomyces pombe) and a decrease in pathogenic fungi, such as Candida [171]. In CDI, the overrepresentation of Candida albicans and reduced fungal diversity are common, while FMT responders tend to gain donor-derived Saccharomyces and Aspergillus species; non-responders, on the other hand, retain high Candida levels [172]. The pre-FMT abundance of Candida spp. in recipients may predict FMT success, and donor feces enriched in fungi, such as Filobasidium, are associated with better outcomes, possibly due to enhanced anti-inflammatory effects [173]. Experimental models further demonstrate that antibiotic- induced dysbiosis promotes fungal overgrowth, which can be suppressed by fecal microbiota transplantation (FMT) highlighting the interplay between bacteria and fungi in colonization resistance [174]. These findings underscore the importance of considering both the virome and mycobiome in FMT. Incorporating their dynamics into future FMT strategies may improve therapeutic outcomes, reduce recurrence, and enable more personalized microbiota-based interventions.
Personalized Medicine Approaches
Personalized medicine is emerging as a powerful strategy for addressing microbiome-related health conditions by tailoring interventions to an individual’s specific microbial and clinical profile. Advances in metagenomics and sequencing technologies now enable comprehensive analysis of the gut microbiota, making it possible for clinicians to design targeted treatments that consider factors such as diet, ancestry, physiology, and microbial signatures [175]. These individualized approaches bridge microbiome research and clinical practice, allowing for precise dietary and therapeutic recommendations based on both host and microbial metabolic profiles [176,177]. A personalized strategy typically involves a detailed analysis of a patient’s clinical data, gut microbiome, and metabolome, identifying shifts in microbial diversity and function. This is crucial given the significant intra-individual variability in microbiota, which can influence therapeutic responses and create drug response biases [178,179]. For example, personalized probiotic regimens based on stool analysis and health history have demonstrated increased beneficial bacterial populations and microbial diversity, resulting in improved outcomes in conditions such as diarrhoea and constipation [180]. As omics data are increasingly integrated into clinical care, advances in bioinformatics and machine learning are essential for interpreting complex datasets. These tools support the development of precision probiotics, next-generation prebiotics, and tailored dietary therapies [181]. Ultimately, the integration of microbiome-based diagnostics and therapeutics with pharmacogenomics and epigenomics marks a new frontier in patient care, moving healthcare toward truly personalized medicine [182].
Conclusion and Future Perspectives
A balanced interplay among gut bacteria, viruses, and fungi is fundamental to maintaining gut homeostasis and overall human health. Disruption of these interactions, through virome or mycobiome dysbiosis, leads to microbial imbalances that contribute to the development of disease. Indeed, shifts in virome or mycobiome composition are linked to a range of conditions, from chronic gut inflammation (e.g., IBD) and autoimmune disorders (such as rheumatoid arthritis) to metabolic diseases (obesity and type 2 diabetes). Such disturbances compromise mucosal barrier integrity, disrupt immune responses, and alter metabolic pathways, leading to both local and systemic effects. Looking ahead, the gut virome and mycobiome remain less well-characterized than the bacteriome, primarily due to persistent technical challenges. Advances in metagenomic sequencing and bioinformatics are beginning to illuminate the vast “viral dark matter” and reveal greater fungal diversity. However, incomplete reference databases and high inter-individual variability still hinder progress. Future studies should prioritize integrated multi-omics approaches to decipher multi-kingdom interactions and their impacts on the host, complemented by targeted experiments to establish causative mechanisms in disease. On the therapeutic front, targeting the virome and mycobiome is a promising strategy. Bacteriophage therapy and microbiome-modulating drugs are being investigated for the treatment of antibiotic-resistant infections and chronic gastrointestinal disorders. Antifungal treatments have shown benefits in alleviating gut inflammation (as in IBD). Additionally, tailored high-fiber diets can beneficially reshape gut microbial communities to improve metabolic outcomes. Ultimately, incorporating virome and mycobiome insights into microbiome-based diagnostics and personalized medicine holds promise for more effective disease prevention and treatment.
Author contribution
All authors have contributed significantly.
Acknowledgement
All authors are thankful to their respective affiliations for the facility’s support in completing this project.
Funding
Not applicable.
Ethics approval
Not applicable.
Consent of Publication
Not applicable.
Competing interest
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, et al. (2014) Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis Nat Med 20(2): 159-166.
- Lee JY, Bays DJ, Savage HP, Baumler AJ (2024) The human gut microbiome in health and disease: time for a new chapter? Infect Immun 92(11): e0030224.
- Grenham S, Clarke G, Cryan JF, Dinan TG (2011) Brain-gut-microbe communication in health and disease. Front Physiol 2: 94.
- Chu H, MazmanianSK (2013) Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol 14(7): 668-675.
- Cabinian A, Sinsimer D, Tang M, Jang Y, Choi B, et al. (2018) Gut symbiotic microbes imprint intestinal immune cells with the innate receptor SLAMF4 which contributes to gut immune protection against enteric pathogens. Gut 67(5): 847-859.
- Cani PD (2018) Human gut microbiome: hopes, threats and promises. Gut 67(9): 1716-1725.
- Dagar S, SinghJ, Saini A, Kumar Y, Chhabra S, et al. (2022) Gut bacteriome, mycobiome and virome alterations in rheumatoid arthritis. Front Endocrinol (Lausanne) 13: 1044673.
- Vemuri R, Shankar EM, Chieppa M, Eri R, Kavanagh K (2020) Beyond Just Bacteria: Functional Biomes in the Gut Ecosystem Including Virome, Mycobiome, Archaeome and Helminths. Microorganisms 8(4): 483.
- Stefanaki C, Peppa M, Mastorakos G, Chrousos GP (2017) Examining the gut bacteriome, virome, and mycobiome in glucose metabolism disorders: Are we on the right track? Metabolism 73: 52-66.
- Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, et al. (2018) Current understanding of the human microbiome. Nat Med 24(4): 392-400.
- Forbes JD, Bernstein CN, Tremlett H, Van Domselaar G, Knox NC (2019) A Fungal World: Could the Gut Mycobiome Be Involved in Neurological Disease? Front Microbiol 9: 3249.
- Mukhopadhya I, Segal JP, Carding SR, Hart AL, Hold GL (2019) The gut virome: the 'missing link' between gut bacteria and host immunity? Therap Adv Gastroenterol 12: 1756284819836620.
- Hoyles L, McCartney AL, Neve H, Gibson GR, Sanderson JD, et al. (2014) Characterization of virus-like particles associated with the human faecal and caecal microbiota. Res Microbiol 165(10): 803-812.
- Carding SR, Davis N, Hoyles L (2017) Review article: the human intestinal virome in health and disease. Aliment Pharmacol Ther 46(9): 800-815.
- Zarate S, Taboada B, Yocupicio-Monroy M, Arias CF (2017) Human Virome. Arch Med Res 48(8): 701-716.
- Matijasic M, Mestrovic T, Paljetak HC, Peric M, Baresic A, et al. (2020) Gut Microbiota beyond Bacteria-Mycobiome, Virome, Archaeome, and Eukaryotic Parasites in IBD. Int J Mol Sci 21(8): 2668.
- Liang G, Bushman FD (2021) The human virome: assembly, composition and host interactions. Nat Rev Microbiol 19(8): 514-527.
- Edwards RA, Vega AA, Norman HM, Ohaeri M, Levi K, et al. (2019) Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat Microbiol 4(10): 1727-1736.
- Song Y, Li X, Wei X, Cui J (2021) Human Endogenous Retroviruses as Biomedicine Markers. Virol Sin 36(5): 852-858.
- Van Espen L, Bak EG, Beller L, Close L, Deboutte W, et al. (2021) A Previously Undescribed Highly Prevalent Phage Identified in a Danish Enteric Virome Catalog. mSystems 6(5): e0038221.
- Pargin E, Roach MJ, Skye A, Papudeshi B, Inglis LK, et al. (2023) The human gut virome: composition, colonization, interactions, and impacts on human health. Front Microbiol 14: 963173.
- McCann A, Ryan FJ, Stockdale SR, Dalmasso M, Blake T, et al. (2018) Viromes of one year old infants reveal the impact of birth mode on microbiome diversity. PeerJ 6: e4694.
- Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, et al. (2016) Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537(7622): 689-693.
- Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, et al. (2015) Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160(3): 447-460.
- Nishida A, Inoue R, Inatomi O, Bamba S, Naito Y, et al. (2018) Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol 11(1): 1-10.
- Chen Y, Cui W, Li X, Yang H (2021) Interaction Between Commensal Bacteria, Immune Response and the Intestinal Barrier in Inflammatory Bowel Disease. Front Immunol 12: 761981.
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, et al. (2014) The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15(3): 382-392.
- Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, et al. (2011) The human gut virome: inter-individual variation and dynamic response to diet. Genome Res 21(10): 1616-1625.
- Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP, et al. (1998) The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol Microbiol 29(2): 527-543.
- Perez-Brocal V, Garcia-Lopez R, Nos P, Beltran B, Moret I, et al. (2015) Metagenomic Analysis of Crohn's Disease Patients Identifies Changes in the Virome and Microbiome Related to Disease Status and Therapy, and Detects Potential Interactions and Biomarkers. Inflamm Bowel Dis 21(11): 2515-2532.
- Khan I, Ullah N, Zha L, Bai Y, Khan A, et al. (2019) Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 8(3): 126.
- Zhao G, Vatanen T, Droit L, Park A, Kostic AD, et al. (2017) Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc Natl Acad Sci U S A 114(30): E6166-E6175.
- Ma Y, You X, Mai G, Tokuyasu T, Liu C (2018) A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome 6(1): 24.
- Meldrum DR, Morris MA, Gambone JC (2017) Obesity pandemic: causes, consequences, and solutions-but do we have the will? Fertil Steril 107(4): 833-839.
- Guo Y, Wang B, Gao H, Gao L, Hua R, et al. (2021) ACE2 in the gut: The center of the 2019-nCoV infected pathology. Frontiers in Molecular Biosciences 8: 708336.
- Krishnamurthy SR, Wang D (2017) Origins and challenges of viral dark matter. Virus Res 239: 136-142.
- Wang D (2020) 5 challenges in understanding the role of the virome in health and disease. PLoS Pathog 16(3): e1008318.
- Yan A, Butcher J, Schramm L, Mack DR, StintziA A (2023) Multiomic spatial analysis reveals a distinct mucosa-associated virome. Gut Microbes 15(1): 2177488.
- Fujimoto K, Miyaoka D, Uematsu S (2022) Characterization of the human gut virome in metabolic and autoimmune diseases. Inflamm Regen 42(1): 32.
- Mirzaei MK, Xue J, Costa R, Ru J, Schulz S, et al. (2021) Challenges of Studying the Human Virome - Relevant Emerging Technologies. Trends Microbiol 29(2): 171-181.
- Benler S, Yutin N, Antipov D, Rayko M, Shmakov S, et al. (2021) Thousands of previously unknown phages discovered in whole-community human gut metagenomes. Microbiome 9(1): 78.
- Li J, Yang F, Xiao M, Li A (2016) Advances and challenges in cataloging the human gut virome. Cell Host Microbe 30(7): 908-916.
- Mottawea W, Chiang CK, Muhlbauer M, Starr AE, Butcher J, Tet al. (2016) Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn's disease. Nat Commun 7: 13419.
- Kootte RS, Levin E, Salojarvi J, Smits LP, Hartstra AV, et al. (2017) Improvement of Insulin Sensitivity after Lean Donor Feces in Metabolic Syndrome Is Driven by Baseline Intestinal Microbiota Composition. Cell Metab 26(4): 611-619 e6.
- Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, et al. (2017) Salt-responsive gut commensal modulates T(H)17 axis and disease. Nature 551(7682): 585-589.
- Baxter NT, Ruffin MT, Rogers MA, Schloss PD (2016) Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med 8(1): 37.
- Zhang X, Shen D, Fang Z, Jie Z, Qiu X, et al. (2013) Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One 8(8): e71108.
- Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, et al. (2015) The Gut Microbiome Contributes to a Substantial Proportion of the Variation in Blood Lipids. Circ Res 117(9): 817-24.
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, et al. (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505(7484): 559-563.
- Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, et al. (2013) Dietary intervention impact on gut microbial gene richness. Nature 500(7464): 585-588.
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, et al. (2005) Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307(5717): 1955-1959.
- Macfarlane S, Macfarlane GT (2003) Regulation of short-chain fatty acid production. Proc Nutr Soc 62(1): 67-72.
- Sartor RB (2008) Microbial influences in inflammatory bowel diseases. Gastroenterology 134(2): 577-94.
- Feitoza AB, Pereira AF, da Costa NF, Ribeiro BG (2009) Conjugated linoleic acid (CLA): effect modulation of body composition and lipid profile. Nutr Hosp 24(4) (2009) 422-428.
- Fukiya S, Arata M, Kawashima H, Yoshida D, Kaneko M, et al. (2009) Conversion of cholic acid and chenodeoxycholic acid into their 7-oxo derivatives by Bacteroides intestinalis AM-1 isolated from human feces. FEMS Microbiol Lett 293(2): 263-270.
- Saha JR, Butler VP, Neu HC, Lindenbaum J (1983) Digoxin-inactivating bacteria: identification in human gut flora. Science 220(4594): 325-327.
- Wallace BD, Wang H, Lane KT, Scott JE, Orans J, et al. (2010) Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330(6005): 831-835.
- Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK (209) Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A 106(34): 14728-14733.
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, et al. (2008) The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105(39): 15064-15069.
- Kim YS, Ho SB (2010) Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep 12(5): 319-3130.
- Podolsky DK, Lynch-Devaney K, Stow JL, Oates P, Murgue B, et al. (1993) Identification of human intestinal trefoil factor. Goblet cell-specific expression of a peptide targeted for apical secretion. J Biol Chem 268(9): 6694-6702.
- Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, et al. (2017) The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5(1): 153.
- Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, et al. (2015) Role of the normal gut microbiota. World J Gastroenterol 21(29): 8787-87803.
- Lutgendorff F, Akkermans LM, Soderholm JD et al. (2008) The role of microbiota and probiotics in stress-induced gastro-intestinal damage. Curr Mol Med 8(4): 282-298.
- Kho ZY, Lal SK (2018) The Human Gut Microbiome - A Potential Controller of Wellness and Disease. Front Microbiol 9: 1835.
- Genth H, Huelsenbeck J, Hartmann B, Hofmann F, Just I, et al. (2006) Cellular stability of Rho-GTPases glucosylated by Clostridium difficile toxin B. FEBS Lett 580(14): 3565-3569.
- Pruitt RN, Chumbler NM, Rutherford SA, Farrow MA, Friedman DB, et al. (2012) Structural determinants of Clostridium difficile toxin A glucosyltransferase activity. J Biol Chem 287(11): 8013-8020.
- Bartlett JG (1981) Antimicrobial agents implicated in Clostridium difficile toxin-associated diarrhea of colitis. Johns Hopkins Med J 149(1): 6-9.
- Lennard-Jones JE (1989) Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl 170: 16-9.
- Loftus EV (2004) Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology 126(6): 1504-1517.
- Donnelly KL, Smith CI, Schwarzenberg SJ, Jessurun J, Boldt MD, et al. (2005) Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Invest 115(5): 1343-1351.
- Peroumal D, Sahu SR, Kumari P, Utkalaja BG, Acharya N (2022) Commensal Fungus Candida albicans Maintains a Long-Term Mutualistic Relationship with the Host To Modulate Gut Microbiota and Metabolism. Microbiol Spectr 10(5): e0246222.
- Sun S, Sun L, Wang K, Qiao S, Zhao X, et al. (2021) The gut commensal fungus, Candida parapsilosis, promotes high fat-diet induced obesity in mice. Commun Biol 4(1): 1220.
- Perez JC (2021) Fungi of the human gut microbiota: Roles and significance. Int J Med Microbiol 311(3): 151490.
- Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, et al. (2015) Activation of HIF-1alpha and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med 21(7): 808-814.
- Underhill DM, Iliev ID (2014) The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol 14(6): 405-416.
- Romani L (2011) Immunity to fungal infections. Nat Rev Immunol 11(4): 275-288.
- Brown GD (2011) Innate antifungal immunity: the key role of phagocytes. Annu Rev Immunol 29: 1-21.
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, ReyesCN, et al. (2012) Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336(6086): 1314-1317.
- Ifrim DC, Bain JM, Reid DM, Oosting M, Verschueren I, et al. (2014) Role of Dectin-2 for host defense against systemic infection with Candida glabrata. Infect Immun 82(3): 1064-1073.
- Netea MG, Graaf CAVD, Vonk AG, Verschueren I, Meer JWVD, et al. (2002) The role of toll-like receptor (TLR) 2 and TLR4 in the host defense against disseminated candidiasis. J Infect Dis 185(10): 1483-1489.
- Stappers MHT, Clark AE, Aimanianda V, Bidula S, Reid DM, et al. (2018) Recognition of DHN-melanin by a C-type lectin receptor is required for immunity to Aspergillus. Nature 555(7696): 382-386.
- Jaeger M, Lee RVD, Cheng SC, Johnson MD, Kumar V, et al. (2015) The RIG-I-like helicase receptor MDA5 (IFIH1) is involved in the host defense against Candida infections. Eur J Clin Microbiol Infect Dis 34(5): 963-974.
- Yeung F, Chen YH, Lin JD, Leung JM, McCauley C, et al. (2020) Altered Immunity of Laboratory Mice in the Natural Environment Is Associated with Fungal Colonization. Cell Host Microbe 27(5): 809-822 e6.
- Bernardes EVT, Pettersen VK, Gutierrez MW, Laforest-Lapointe I, et al. (2020) Intestinal fungi are causally implicated in microbiome assembly and immune development in mice. Nat Commun 11(1): 2577.
- Li XV, Leonardi I, Iliev ID (2019) Gut Mycobiota in Immunity and Inflammatory Disease. Immunity 50(6): 1365-1379.
- Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, et al. (2017) Fungal microbiota dysbiosis in IBD. Gut 66(6): 1039-1048.
- Wetzel S, Muller A, Kohnert E, Mehrbarzin N, Huber R, et al. (2023) Longitudinal dynamics of gut bacteriome and mycobiome interactions pre- and post-visceral surgery in Crohn's disease. Front Cell Infect Microbiol 13: 1275405.
- Kotlowski R, Bernstein CN, Sepehri S, Krause DO (2007) High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 56(5): 669-675.
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, et al. (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 104(34) (2007) 13780-13785.
- Cao Y, Oh J, Xue M, Huh WJ, Wang J, et al. (2022) Commensal microbiota from patients with inflammatory bowel disease produce genotoxic metabolites, Science 378(6618) eabm3233.
- Zheng L, Wen XL, Duan SL (2022) Role of metabolites derived from gut microbiota in inflammatory bowel disease, World J Clin Cases 10(9): 2660-2677.
- Canavan C, West J, Card T (2014) Review article: the economic impact of the irritable bowel syndrome, Aliment Pharmacol Ther 40(9): 1023-34.
- El-Serag HB, Pilgrim P, Schoenfeld P (2004) Systemic review: Natural history of irritable bowel syndrome, Aliment Pharmacol Ther 19(8): 861-70.
- Engsbro AL, Simren M, Bytzer P (2012) Short-term stability of subtypes in the irritable bowel syndrome: prospective evaluation using the Rome III classification, Aliment Pharmacol Ther 35(3): 350-9.
- Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, et al. (2017) Global patterns and trends in colorectal cancer incidence and mortality, Gut 66(4): 683-691.
- Li Y, Kundu P, Seow SW, de Matos CT, Aronsson L, Chin KC, et al. (2012) Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APCMin/+ mice, Carcinogenesis 33(6): 1231-8.
- Pandey H, Tang DWT, Wong SH, Lal D(2023) Gut Microbiota in Colorectal Cancer: Biological Role and Therapeutic Opportunities, Cancers (Basel) 15(3): 866.
- Richard ML, Liguori G, Lamas B, Brandi G, da Costa G, et al.(2018) Mucosa-associated microbiota dysbiosis in colitis associated cancer, Gut Microbes 9(2): 131-142.
- Liu YX, Qin Y, Chen T, Lu M, Qian X, et al. (2021) A practical guide to amplicon and metagenomic analysis of microbiome data, Protein Cell 12(5): 315-330.
- Hallen-Adams HE, Suhr MJ (2017) Fungi in the healthy human gastrointestinal tract, Virulence 8(3): 352-358.
- Galloway-Pena J, Iliev ID, McAllister F (2024) Fungi in cancer, Nat Rev Cancer 24(5): 295-298.
- Saftien A, Puschhof J, Elinav E (2023) Fungi and cancer, Gut 72(7): 1410-1425.
- Narunsky-Haziza L, Sepich-Poore GD, Livyatan I, Asraf O, Martino C, et al(2022) Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions, Cell 185(20): 3789-3806 e17.
- Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, et al. (2019) Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer, Gut 68(4): 654-662.
- Lin Y, Lau HC, Liu Y, Kang X , Wang Y, et al. (2022) Altered Mycobiota Signatures and Enriched Pathogenic Aspergillus rambellii Are Associated With Colorectal Cancer Based on Multicohort Fecal Metagenomic Analyses, Gastroenterology 163(4): 908-921.
- Jansen D, Matthijnssens J (2023) The Emerging Role of the Gut Virome in Health and Inflammatory Bowel Disease: Challenges, Covariates and a Viral Imbalance, Viruses 15(1): 173.
- Ungaro F, Massimino L, D'Alessio S, Danese S (2019) The gut virome in inflammatory bowel disease pathogenesis: From metagenomics to novel therapeutic approaches, United European Gastroenterol J 7(8): 999-1007.
- Hsu C, Ghannoum M, Cominelli F, Martino LD (2023) Mycobiome and Inflammatory Bowel Disease: Role in Disease Pathogenesis, Current Approaches and Novel Nutritional-based Therapies, Inflamm Bowel Dis 29(3): 470-479.
- Fang J, Xie C, Tao Y, Wei D (2023) An overview of single-molecule techniques and applications in the study of nucleic acid structure and function, Biochimie 206: 1-11.
- Kiseleva O, Ponomarenko E, Poverennaya E (2020) Empowering Shotgun Mass Spectrometry with 2DE: A HepG2 Study, Int J Mol Sci 21(11): 3813.
- Dacre J, Colligan M, Clarke T, Ammer JJ, Schiemann J, et al. (2021) A cerebellar-thalamocortical pathway drives behavioral context-dependent movement initiation, Neuron 109(14): 2326-2338 e8.
- Chen C, Yan Q, Yao X, Li S, Lv Q, et al. (2022) Alterations of the gut virome in patients with systemic lupus erythematosus, Front Immunol 13: 1050895.
- Martinez-Medina M, Garcia-Gil LJ (2014) Escherichia coli in chronic inflammatory bowel diseases: An update on adherent invasive Escherichia coli pathogenicity, World J Gastrointest Pathophysiol 5(3): 213-27.
- Turnbull AD, Guerra J, Starnes HF (1989) Results of surgery for obstructing carcinomatosis of gastrointestinal, pancreatic, or biliary origin, J Clin Oncol 7(3): 381-6.
- Zhang L, Zhan H, Xu W, Yan S, Ng SC (2021) The role of gut mycobiome in health and diseases, Therap Adv Gastroenterol 14: 17562848211047130.
- Fang L, Ning J (2024) Gut virome and diabetes: discovering links, exploring therapies, Arch Microbiol 206(8): 346.
- Daliri EB, Ofosu FK, Chelliah R, Lee BH, Oh DH (2020) Health Impact and Therapeutic Manipulation of the Gut Microbiome, High Throughput 9(3): 17.
- Benjamin A, Sultan A, Yousif M, Moussa A, Abdo EF, et al. (2022) Qualitative healthcare worker survey: Retrospective cross-sectional case study on COVID-19 in the African context, Ann Med Surg (Lond) 79: 103918.
- Liang G, Cobian-Guemes AG, Albenberg L, Bushman F (2021) The gut virome in inflammatory bowel diseases, Curr Opin Virol 51: 190-198.
- Wylie KM, Weinstock GM, Storch GA (2012) Emerging view of the human virome, Translational research 160(4): 283-290.
- Pollock J, Glendinning L, Wisedchanwet T, Watson M (2018) The madness of microbiome: attempting to find consensus “best practice” for 16S microbiome studies, Applied and environmental microbiology 84(7): e02627-17.
- Cao Z, Sugimura N, Burgermeister E, Ebert MP, Zuo T, et al. (2022) The gut virome: A new microbiome component in health and disease, EBioMedicine 81.
- Santiago-Rodriguez TM, Hollister EB (2019) Human virome and disease: high-throughput sequencing for virus discovery, identification of phage-bacteria dysbiosis and development of therapeutic approaches with emphasis on the human gut, Viruses 11(7): 656.
- Raeisi H, Noori M, Azimirad M, Mohebbi SR, Asadzadeh Aghdaei H, et al. (2023) Emerging applications of phage therapy and fecal virome transplantation for treatment of Clostridioides difficile infection: challenges and perspectives, Gut pathogens 15(1): 21.
- Acevedo-Román A, Pagán-Zayas N, Velázquez-Rivera LI, Torres-Ventura AC, Godoy-Vitorino F (2024) Insights into gut dysbiosis: inflammatory diseases, obesity, and restoration approaches, International Journal of Molecular Sciences 25(17): 9715.
- Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ (2015) Dysbiosis of the gut microbiota in disease, Microbial ecology in health and disease 26(1): 26191.
- Mahdavi-Roshan M, Salari A, Kheirkhah J, Ghorbani Z (2022) The effects of probiotics on inflammation, endothelial dysfunction, and atherosclerosis progression: a mechanistic overview, Heart, Lung and Circulation 31(5): e45-e71.
- Tyszkowski R, Mehrzad R (2023) Inflammation: a multifaceted and omnipresent phenomenon, Inflammation and Obesity, Elsevier pp19-30.
- Strathdee, Hatfull GF, Mutalik VK (2023) Schooley, Phage therapy: From biological mechanisms to future directions, Cell 186(1): 17-31.
- Kapoor A, Mudaliar SB, Bhat VG, Chakraborty I, Prasad ASB, et al. (2024) Phage therapy: A novel approach against multidrug-resistant pathogens, 3 Biotech 14(10): 256.
- Gordillo Altamirano FL, Barr JJ (2019) Phage therapy in the postantibiotic era, Clinical microbiology reviews 32(2): 10.1128/cmr. 00066-18.
- Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GG, et al. (2014) A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes, Nature communications 5(1): 4498.
- Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, et al. (2016) Healthy human gut phageome, Proceedings of the National Academy of Sciences 113(37): 10400-10405.
- Maronek M, Link R, Ambro L, Gardlik R (2020) Phages and their role in gastrointestinal disease: focus on inflammatory bowel disease, Cells 9(4): 1013.
- Moelling K, Broecker F, Willy C (2018) A wake-up call: we need phage therapy now, Viruses 10(12): 688.
- Rea K, Dinan TG, Cryan JF (2016) The microbiome: A key regulator of stress and neuroinflammation, Neurobiology of stress 4: 23-33.
- Lin DM, Koskella B, Lin HC (2017) Phage therapy: An alternative to antibiotics in the age of multi-drug resistance, World journal of gastrointestinal pharmacology and therapeutics 8(3): 162.
- Federici S, Kredo-Russo S, Valdés-Mas R, Kviatcovsky D, Weinstock E, et al. (2022) Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation, Cell 185(16): 2879-2898. e24.
- Paule A, Frezza D, Edeas M (2018) Microbiota and phage therapy: future challenges in medicine, Medical sciences 6(4): 86.
- Hu YO, Hugerth LW, Bengtsson C, Alisjahbana A, Seifert M, et al. (2018) Bacteriophages synergize with the gut microbial community to combat Salmonella, MSystems 3(5): 10.1128/msystems. 00119-18.
- Chaudhry WN, Concepcion-Acevedo J, Park T, Andleeb S, Bull JJ, et al. (2017) Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms, PloS one 12(1): e0168615.
- Sarker SA, Sultana S, Reuteler G, Moine D, Descombes P, et al. (2016) Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh, EBioMedicine 4: 124-137.
- Gindin M, Febvre HP, Rao S, Wallace TC, Weir TL (2019)Bacteriophage for gastrointestinal health (PHAGE) study: evaluating the safety and tolerability of supplemental bacteriophage consumption, Journal of the American College of Nutrition 38(1): 68-75.
- Febvre HP, Rao S, Gindin M, Goodwin ND, Finer E, et al. (2019) PHAGE study: effects of supplemental bacteriophage intake on inflammation and gut microbiota in healthy adults, Nutrients 11(3): 666.
- Bach S, Johnson R, Stanford K, McAllister T (2009) Bacteriophages reduce Escherichia coli O157: H7 levels in experimentally inoculated sheep, Canadian journal of animal science 89(2): 285-293.
- Hosseinidoust Z (2019) Phage Therapy with a Focus on the Human Microbiota, Antibiotics (Basel, Switzerland) 8(3): E131-E131.
- Titécat M, Rousseaux C, Dubuquoy C, Foligné B, Rahmouni O, et al. (2022) Safety and efficacy of an AIEC-targeted bacteriophage cocktail in a mice colitis model, Journal of Crohn's and Colitis 16(10): 1617-1627.
- Pinchera B, Scotto R, Zappulo E, Buonomo AR, Maraolo AE, et al. (2023) Impact of oral antiviral therapy against HCV on gut microbiota. A prospective study, The New Microbiologica 46(2): 196-201.
- Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, et al. (2015) Disease-specific alterations in the enteric virome in inflammatory bowel disease, Cell 160(3): 447-460.
- Lockhart A, Mucida D, Parsa R (2022) Immunity to enteric viruses, Immunity 55(5): 800-818.
- Sundararaman A, Ray M, Ravindra P, Halami PM (2020) Role of probiotics to combat viral infections with emphasis on COVID-19, Applied microbiology and biotechnology 104: 8089-8104.
- Gáspár Z, Nagavci B, Szabó BG, Lakatos B (2024) Gut Microbiome Alteration in HIV/AIDS and the role of antiretroviral Therapy- A scoping review, Microorganisms 12(11): 2221.
- Lee H, Ko G (2017) New perspectives regarding the antiviral effect of vitamin A on norovirus using modulation of gut microbiota, Gut Microbes 8(6): 616-620.
- El Jaddaoui I, Sehli S, Al Idrissi N, Bakri Y, Belyamani L, et al. (2025) The gut mycobiome for precision medicine, Journal of Fungi 11(4): 279.
- Huang H, Wang Q, Yang Y, Zhong W, He F, et al. (2024) The mycobiome as integral part of the gut microbiome: crucial role of symbiotic fungi in health and disease, Gut Microbes 16(1): 2440111.
- Rayens E, Norris KA (2022) Prevalence and healthcare burden of fungal infections in the United States, 2018, Open forum infectious diseases, Oxford University Press US 9(1): ofab593.
- de Souza CM, Bezerra BT, Mellon DA, de Oliveira HC (2025) The evolution of antifungal therapy: Traditional agents, current challenges and future perspectives, Current Research in Microbial Sciences 100341.
- Wu Y, Hu S, Wu C, Gu F, Yang Y (2022) Probiotics: potential novel therapeutics against fungal infections, Frontiers in Cellular and Infection Microbiology 11: 793419.
- Divyashree S, Shruthi B, Vanitha P, Sreenivasa M (2023) Probiotics and their postbiotics for the control of opportunistic fungal pathogens: A review, Biotechnology Reports 38: e00800.
- Guo J, Brosnan B, Furey A, Arendt E, Murphy P, et al. (2012) Antifungal activity of Lactobacillus against Microsporum canis, Microsporum gypseum and Epidermophyton floccosum, Bioengineered 3(2): 104-113.
- Ajah HA (2016) In vitro and in vivo studies on the antifungal activity of probiotics and seaweed extract (Ascophyllum nodosum), Int. J. Innov. Sci. Eng. Technol 3: 306-312.
- Matsubara VH, Bandara H, Mayer MP, Samaranayake LP (2016) Probiotics as antifungals in mucosal candidiasis, Clinical Infectious Diseases 62(9): 1143-1153.
- Romeo M, Romeo D, Trovato L, Oliveri S, Palermo F, et al. (2011) Role of probiotics in the prevention of the enteric colonization by Candida in preterm newborns: incidence of late-onset sepsis and neurological outcome, Journal of Perinatology 31(1): 63-69.
- Roy A, Chaudhuri J, Sarkar D, Ghosh P, Chakraborty S (2014) Role of enteric supplementation of probiotics on late-onset sepsis by Candida species in preterm low birth weight neonates: a randomized, double blind, placebo-controlled trial, North American journal of medical sciences 6(1): 50.
- Lam S, Bai X, Shkoporov AN, Park H, Wu X, et al. (2022) Roles of the gut virome and mycobiome in faecal microbiota transplantation, The lancet Gastroenterology & hepatology 7(5): 472-484.
- Zuo T, Wong SH, Lam K, Lui R, Cheung K, et al. (2018) Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome, Gut 67(4): 634-643.
- Draper LA, Ryan FJ, Smith MK, Jalanka J, Mattila E, et al. (2018) Long-term colonisation with donor bacteriophages following successful faecal microbial transplantation, Microbiome 6: 1-9.
- Rasmussen TS, Koefoed AK, Jakobsen RR, Deng L, Castro-Mejía JL, et al. (2020) Bacteriophage-mediated manipulation of the gut microbiome–promises and presents limitations, FEMS Microbiology Reviews 44(4): 507-521.
- Draper LA, Ryan FJ, Dalmasso M, Casey PG, McCann A, et al. (2020) Autochthonous faecal viral transfer (FVT) impacts the murine microbiome after antibiotic perturbation, BMC biology 18: 1-14.
- Chen Q, Fan Y, Zhang B, Yan C, Chen Z, et al. (2023) Specific fungi associated with response to capsulized fecal microbiota transplantation in patients with active ulcerative colitis, Frontiers in Cellular and Infection Microbiology 12: 1086885.
- Zuo T, Wong SH, Cheung CP, Lam K, Lui R, et al. (2018) Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection, Nature communications 9(1): 3663.
- van Thiel IA, Rahman S, Hakvoort TB, Davids M, Verseijden C, et al. (2022) Fecal filobasidium is associated with clinical remission and endoscopic response following fecal microbiota transplantation in mild-to-moderate ulcerative colitis, Microorganisms 10(4): 737.
- Matsuo K, Haku A, Bi B, Takahashi H, Kamada N, et al. (2019) Fecal microbiota transplantation prevents Candida albicans from colonizing the gastrointestinal tract, Microbiology and immunology 63(5): 155-163.
- Shukla V, Singh S, Verma S, Verma S, Rizvi AA, et al. (2024) Targeting the microbiome to improve human health with the approach of personalized medicine: latest aspects and current updates, Clinical Nutrition ESPEN 63: 813-820.
- Jain N (2020) The need for personalized approaches to microbiome modulation, Frontiers in public health 8: 144.
- Abeltino A, Hatem D, Serantoni C, Riente A, De Giulio MM, et al. (2024) Unraveling the gut microbiota: implications for precision nutrition and personalized medicine, Nutrients 16(22): 3806.
- Bomba A, Haranta M (2023) Personalized and Targeted Gut Microbiome Modulation in the Prevention and Treatment of Chronic Diseases, Advances in Probiotics for Health and Nutrition, IntechOpen.
- Huang G, Khan R, Zheng Y, Lee PC, Li Q, et al. (2023) Exploring the role of gut microbiota in advancing personalized medicine, Frontiers in Microbiology 14: 1274925.
- Ryšávka P, Kohutková-Lánová M, Netušil R, Pospíšilová E, Hanáčková M, et al. (2022) Targeted Modulation of Gut Microbiota by Personalized Probiotics, International Journal of Probiotics & Prebiotics 17(1).
- Wu J, Singleton SS, Bhuiyan U, Krammer L, Mazumder R (2024) Multi-omics approaches to studying gastrointestinal microbiome in the context of precision medicine and machine learning, Frontiers in molecular biosciences 10: 1337373.
- Kashyap PC, Chia N, Nelson H, Segal E, Elinav E (2017) Microbiome at the frontier of personalized medicine, Mayo Clinic Proceedings, Elsevier 92(12): 1855-1864.






























