Characterizing the Nitroproteome Using Bioinformatic Tools: A Mini-Review
Swarnab Sengupta and Arindam Bhattacharjee*
Department of Microbiology, University of North Bengal, India
Submission: February 06, 2017; Published: March 23, 2017
*Corresponding author: Arindam Bhattacharjee, Assistant Professor, Department of Microbiology, University of North Bengal, Raja Rammohunpur, Siliguri, Darjeeling, West Bengal, pin: 734013, India, Tel: +919674665722; Email: aribiochem@gmail.com
*How to cite this article: Swarnab S, Arindam B. Characterizing the Nitroproteome Using Bioinformatic Tools: A Mini-Review. Biostat Biometrics Open Acc J. 2017;1(1): 555553. DOI: 10.19080/BBOAJ.2017.01.555553
Abstract
Protein tyrosine nitration (PTN) is a post-translational modification that occurs due to the action of reactive nitrogen species (RNS). The specific tyrosine residues are mainly nitrated in the presence of nitrating agents. It is also related to neurodegenerative diseases, cellular signaling. So the characterization of nitroproteome with their functions is very much essential. In recent times bioinformatic tools become a powerful apparatus to characterize the biological objects. In this mini-review we have attempted to give an outline about the bioinformatic tools which can be used to characterize the nitration/denitration system.
Keywords: Protein tyrosine nitration; Reactive nitrogen species; Nitroproteome; Bioinformatic tools
Abbreviations: RNS: Reactive Nitrogen Species; NO2-TYR: 3-Nitrotyrosine; PTN: Protein Tyrosine Nitration
Introduction
Nitrosative stress is a condition in which the cellular redox homoeostasis is changed because of excessive production of different reactive nitrogen species (RNS) [1]. Under nitrosative stress the ratio of nitrosants to antioxidants is always >1. One of the major markers of nitrosative stress is the 3-Nitrotyrosine (NO2-TYR), a stable post-translational modification of protein. 3-Nitrotyrosine forms due to the reaction of tyrosine and nitrating agents. A nitro (-NO2) group is added in the ortho position of the phenolic hydroxyl group of tyrosine during the reaction of tyrosine and nitrating agent which results in the formation of 3-Nitrotyrosine (NO2-TYR). Generally the natural abundance of tyrosine residues is about 3% in the proteins. But the nitration may introduce negative charge at neutral pH which may results in the change of the local physiological and chemical environment of the biomolecules. Due to this event the structure and function of the proteins are also altered. Thus, the cellular mechanism is also changed. 3-Nitrotyrosine has an impact on clinical biology [2]. It is related with the cell signaling and disease initiation and progression like neurodegenerative diseases, cancer and cardiovascular injury. Alzheimer’s disease, the most common neurodegenerative disease is associated with the formation of 3-Nitrotyrosine. Alzheimer’s disease is induced by accumulation of nitrated tau and mis folded ap proteins in the brain [3]. It is also reported that proteins like alcholo dehydrogenase [4], aldolase [5], isocitrtae dehydrogenase [6] can be nitrated during nitrosative stress. Some enzymes are also present to counteract the nirosative insult e.g. catalase [7],Cytochorme C [8] etc. but there are some reports that suggest these stress response enzymes can also be nitrated in the presence of nitrating agents. Interestingly, some reports suggest that PTN is also related to cellular signaling. The proteins of the mating signaling pathway can be nitrated in Saccharomyces cerevisiae [9]. So this event gives an idea about denitration. But the harmony between the balance of nitration and denitration is yet to be elucidated.
Thus, it is assumed that the nitration/de nitration pathway is just phosphorylation/dephosphorylation. So right now one of the major challenges for biologist is to characterize the nitration/denitration path way. For this purpose one of the most important tool is bioinformatic. In this study we have tried to give an outline about the bioinformatics tools which can be used to understand the biochemistry of protein tyrosine nitration and de nitration.
Bioinformatic tools
Bioinformatic tools are used in the proteomics study not only to characterize the nitroproteins, their structure and function but this specific [10]; sensitive process is also related to the secondary structure of protein, Solvent accessibility [11], and protein-protein interactions [12]. The study of protein tyrosine nitration/denitration is started from the identification of nitroproteins. The mostly used software to identify the nitropoteins is Turbo SEQUEST [10]. One of the important software is Scaffold software which helps to compute protein and peptide probability for nitration [13]. MS-BLAST is also applied to identify the homolog's of different nitro proteins which needs de novo correct tagging. To ensure the correct tagging SPIDER software is used where de novo sequencing and homology mutations are taken into account [14].
The secondary structure of protein is needed to be characterized to identify the nitration-prone tyrosine residues. PSIPRED (Software for Protein Identification from Sequence Tags with De Novo Sequencing Error), JNET, PROF Etc. [15-19]. Algorithms are used to characterize the secondary structure. Neutral networking is one of the best methods to characterize the secondary structure. JPred4 is the latest version of JPred to predict the secondary structure of proteins. JNet algorithm is used in JPred4 software. JNet algorithm is an error-free method which results in the higher accuracy of JPred 4 [20]. Useful software for characterizing the PTN site is GPS-YNO2 having high accuracy [21]. Solvent accessibility another important factor to characterize the protein structure is efficiently predicted by SANN, a nearest neighbor method [22].
Protein networking, another important bioinformatics method used to determine the role and interaction of a protein in certain protein pool. PIN (protein interaction network) and PSN (Protein-Signaling Network) are two major network models for proteomics. The post translational modifications are actually studied by PSN whereas PIN is based on the proteinprotein bindings [12]. The databases for protein interactions are DIP, BIND, MIPS, and MINT etc. One of the strongest software JDIP is used in DIP data model which consists of binary protein interactions. The information about the interacting proteins is also got from DIP data model [23]. DIP is mainly used for budding yeast because a huge amount of external information for budding yeast is only available now [24]. One of the largest collections of freely available information about pair wise molecular interactions and complexes is BIND (Bimolecular Interaction Network Database). The interaction between biological objects like DNA, RNA, gene, protein is characterized by BIND. The specification of BIND data is available as XML DTD and ASN.1 [25]. The indirect and genetic interaction is predicted by MINT (The Molecular Interaction database). Information regarding the modification of enzymes, their kinetics and their binding domains are stored by MINT [26]. The dataset of high- quality experimental protein-protein interaction in mammals is MIPS mammalian protein-protein interaction database (MPPI)[27].The increasing data content is a problem for visualization. Cytoscape, stronger freely available, open-source java-based network visualization and analysis tool is used for visualization[28].
These softwares may be helpful to characterize the protein tyrosine nitration and denitration system. Protein networking is the choice of bioinformatic tool to establish the probable pathway because it is hypothesized that several enzymes are involved in denitration system. Prediction of secondary structure and Solvent accessibility are also very important to predict the structure and stability of the protein. The probable model for nitration/denitration is outlined in Figure 1.
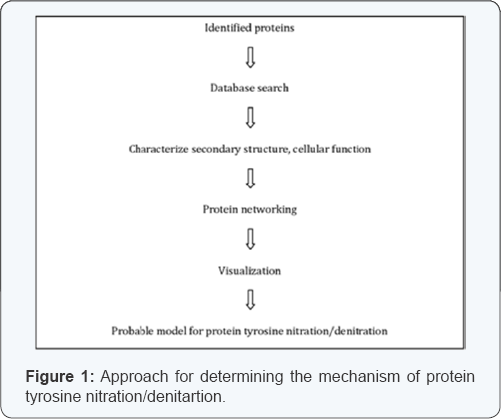
Conclusion
PTN is a topic of ongoing research. PTN has the drastic effect on neurodegenerative diseases. Nitration of neuroproteins results in the tangle formation. The available medicine for these diseases is very few and having side effects. So it is very important to characterize the denitration system. Denitration is the mechanism which can reverse back the nitration, this reaction results in the prevention of tangle formation. So "denitrase" enzyme can be used as the therapeutic medicine for neurodegenerative diseases. But still a huge area of research is needed to understand the nitration/denitration properly
Acknowledgement
The authors acknowledge the University of North Bengal for providing essential infrastructure to carry out this research.
References
- Klatt P Lamas S (2000) Regulation of protein function by S- glutathiolation in response to oxidative and nitrosative stress. Eur J Biochem 267(16): 4928-4944.
- Frears ER, Zhang Z, Blake DR, O'connell JP, Winyard PG (1996) Inactivation of tissue inhibitor metalloproteinase-1 by peroxynitrite. FEBS Lett 381(1-2): 21-24.
- Hashimoto M, Rockenstein E, Crews L, Masliah E (2003) Role of protein aggregation in mitochondrial dysfunction and neurodegeneration in Alzheimer's and Parkinson's diseases. Neuromolecular Med 4(1-2): 21-36.
- Sekar Y, Moon TC, Slupsky CM, Befus AD (2010) Protein Tyrosine Nitration of Aldolase in Mast Cells: A Plausible Pathway in Nitric Oxide- Mediated Regulation of Mast Cell Function. J Immunol 185(1): 578-587.
- Crow JP, Beckman JS, McCord JM (1995) Sensitivity of the Essential Zinc-Thiolate Moiety of Yeast Alcohol Dehydrogenase to Hypochlorite and Peroxynitrite? Biochemistry 34(11): 3544-3552.
- Bhattacharjee A, Majumdar U, Maity D, Sarkar TS, Goswami AM, et al. (2009) In vivo protein tyrosine nitration in S. cerevisiae: identification of tyrosine-nitrated proteins in mitochondria. Biochem Biophys Res Commun 388(3): 612-617.
- Sahoo R, Bhattacharjee A, Majumdar U, Ray SS, Dutta T, et al. (2009) A novel role of catalase in detoxification of peroxynitrite in S. cerevisiae. Biochem Biophys Res Commun 385(4): 507-511.
- Cross R, Aish J, Paston SJ, Poole RK, Moir JW (2000) Cytochrome from Rhodobacter capsulatus confers increased resistance to nitric oxide. J Bacteriol 182(5): 1442-1447.
- Kang JW, Lee NY, Cho KC, Lee MY, Choi DY, et al. (2015) Analysis of nitrated proteins in Saccharomyces cerevisiae involved in mating signal transduction. Proteomics 15(2-3): 580-590.
- Lundgren DH, Han DK, Eng JK (2005) Protein identification using TurboSEQUEST. Curr Protoc Bioinformatics. 13: 13.3.
- Yeo WS, Kim YJ, Kabir MH, Kang JW, KP Kim (2015) Mass spectrometric analysis of protein tyrosine nitration in aging and neurodegenerative diseases. Mass Spectrom Rev 34(2): 166-183.
- Pieroni E, Bentem SF, Mancosu G, Capobianco E, Hirt H, et al. (2008) Protein networking: insights into global functional organization of proteomes. Proteomics 8(4): 799-816.
- Searle BC (2010) Scaffold: a bioinformatic tool for validating MS/MS- based proteomic studies. Proteomics 10(6): 1265-1269.
- Han Y, Ma B, Zhang K (2005) SPIDER: Software for Protein Identification from Sequence Tags with De Novo Sequencing Error. J Bioinform Comput Biol 3(3): 697-716.
- Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292(2): 195-202.
- McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16(4): 404-405.
- Cuff JA, Barton GJ (2000) Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins 40(3): 502-511.
- Ouali M, King RD (2000) Cascaded multiple classifiers for secondary structure prediction. Protein Sci 9(6): 1162-1176.
- Rost B (2001) Review: protein secondary structure prediction continues to rise. J Struct Biol 134(2-3): 204-218.
- Drozdetskiy A, Cole C, Procter J, Barton G J (2015) JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43(W1): W389-W394.
- Liu Z, Cao J, Ma Q, Gao X, Ren J, et al. (2011) GPS-YNO2: computational prediction of tyrosine nitration sites in proteins. Mol Biosyst 7(4): 1197-1204.
- Joo K, Lee SJ, Lee J (2012) Sann: solvent accessibility prediction of proteins by nearest neighbor method. Proteins 80(7): 1791-1797.
- Xenarios I, Salwinski L, Duan XJ, Higney P, Kim S, et al. (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res 30(1): 303-305.
- Deane CM, Salwinski L, Xenarios I, Eisenberg D (2002) Protein Interactions, two methods for assessment of the reliability of high throughput observations. Mol Cell Proteomics 1(5): 349-356.
- Bader GD, Betel D, Hogue CWV (2003) BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res 31(1): 248-250.
- Zanzoni A, Montecchi Palazzi L, Quondam M, Ausiello G, Helmer Citterich M, et al. (2002) MINT: a Molecular INTeraction database. FEBS Lett 513(1): 135-140.
- Pagel P, Kovac S, Oesterheld M, Brauner B, Dunger Kaltenbach I, et al. (2005) The MIPS mammalian protein -protein interaction database. Bioinformatics 21(6): 832-834.
- Smoot ME, Ono K, Ruscheinski J, Wang P, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27(3): 431-432.






























