Variability Heritability and Genetic Advance of Some Groundnut Genotypes (Arachis Hypogaea L.) under Saline Sodic Soil
Omima B HajHussein1*, Ahmed H Abu Assar2, Adel M Fraah1 and Amin Al Sir A3
1Soba Agricultural Research Station, Sudan
2Gezira Agricultural Research Station, Sudan
3Damazin Agricultural Research Station, Sudan
Submission: December 09, 2017; Published: January 08, 2018
*Corresponding author: Omima B HajHussein, Soba Agricultural Research Station, P.O. Box 126 Khartoum, Sudan, Email: aminalnosh@yahoo.com
How to cite this article: Omima B H H, Ahmed H A A, Adel M F, Amin A S A.Variability Heritability and Genetic Advance of Some Groundnut Genotypes (Arachis Hypogaea L.) under Saline Sodic Soil. Ann Rev Resear. 2018; 1(1): 555554. DOI: 10.19080/ARR.2018.01.555554
Abstract
An investigation was carried out at the demonstration farm of Soba research station for reclamation of saline and sodic soils, Khartoum, Sudan, during three successive growing seasons of (2012/13-2013/14-2014/15). The trail was conducted in a randomized block design with three replications to study genetic variability, heritability and genetic advance of yield and yield contributing characters in eight groundnut genotypes under saline sodic soil. Highly significant variations were observed for all characters under studies. High genotypic and phenotypic coefficients of variation were observed for all characters except days to 50% flowering, plant high, pod yield/ha and seed yield/ha. High heritability estimate coupled with high genetic advance were obtained with number of pod/plant, number of seed/plant, pod yield/plant, seed yield/plant and 100 seed mass.
Keywords: Variability; Heritability; Genetic Advance; Groundnut; Saline Sodic Soil
Introduction
Salinity is day after day become a great increasing problem and represents challenge for today's agriculture; it's one of the important a biotic stresses limiting crop production in arid and semi-arid regions, where soil salt content is naturally high and precipitation can be insufficient for leaching [1,2]. Salt-affected soils in Sudan occur in desert, semi- desert climatic zones and in arid regions [3].
Groundnut Arachis hypogaea L is an important crop grown in an area of about 26 million ha, in more than 100 countries around the world, under different agro-climatic conditions for its nutritious oil rich seeds [4], As other plants, groundnut is affected by salinity in all of its developmental stages: from germination to yield [4-10].
Increasing population leads to intensive use of land and water, so it is necessary to bring marginal resources under use. Soil amelioration against salinity is a costly and time-consuming procedure; hence, the alternative strategy is to develop salinity- tolerant genotypes [11]. At the genetic level, salinity tolerance has been considered to be a quantitative trait [12-14], Information on the nature and magnitude of variability and heritability is one of the prerequisites for a successful breeding program in selected genotypes with desirable characters [15]. It is, therefore, of great importance for breeders to know the heritability of the agronomic characters to improve the yield of the crop effectively. According to Falconer & Mackay [16], heritability is defined as the measure of the correspondence between breeding value and phenotypic value. Thus, heritability plays a predictive role in breeding, expressing the reliability of phenotype as a guide to its breeding value. There is a direct relationship between heritability and response to selection, which is referred to as genetic advance. High genetic advance with high heritability estimates offer the most effective condition for selection. The utility of heritability therefore increase when it is used to calculate of genetic advance, which indicates the degree of gain in a character, obtained under a particular selection pressure. Thus genetic advance is another important selection parameter that aids breeder in a selection program.
In Sudan, groundnut is considered one of the most important cash and edible oil crops. Sudan is one of top ten producing countries of groundnut. In spite of this importance, little is known about the effect of salinity on groundnut, and there are no attempts to develop salinity tolerant genotypes. The present study was undertaken to achieve the following objectives:
a) To estimate variability for yield and yield components among some groundnut genotypes under saline soil conditions.
b) To estimate heritability and genetic advance for yield and yield components among some groundnut genotypes under saline soil conditions.
c) To put guide line to establish breeding program for developing genotype (s) tolerant to salinity through identify the suitable selection criteria for higher pod yield through study of variability under saline condition.
Materials and Methods
This study was conducted on the demonstration farm of Soba research station for reclamation of saline and sodic soils, Khartoum, Sudan (EC=4.2dS/m, PH=8.6 and SAR=23) during three successive growing seasons of (2012/13 -2013/14 -2014/15). The plant materials for this investigation comprised eight groundnut cultivars all were released by ARC, Sudan namely; Ahmadi, Bunting, Keriz, Madani, MH383 and Tozi (released for irrigation condition) in addition to, Gebash and Sodari (released for rain fed condition). The experiment was laid out in a Randomized Block Design (RBC) with 3 replicates. Each genotypes in a replicate was grown in a plot of a growth area of 4x5m2. The inter-row and intra-row spacing were 20x70 cm respectively. The recommended packages practices for cultivation of groundnut under irrigation were adopted. Data recorded consist of: days to 50% flowering, plant height (cm), number of branches/plant, number of pods/plant, number of seeds/plant, pods yield/plant (g), seeds yield/plant (g), 100- seed weight (g), pods yield/ha (kg) and seeds yield/ha (kg). Data were statistically analyzed by combine ANOVA and means were separated by Fisher's protected least significant differences (LSD) at p< 0.01 level according to Gomez and Gomez [17].
In order to assess and quantify the genetic variability among the genotypes for the characters under investigation the variance components and values of heritability and genetic advance were estimated following the formula suggested by Burton & De Vane [18] & Johnson et al. [19]. Phenotypic and genotypic variances were estimated using the following formula:
δ2g= (MS g-MS g x y) / r y
δ2g x y = (MS g x y-MS e)/ r
δ = MS e
δ = δ + δ2 g x y / y + δ / r y
Where, δ2g = genotypic variance; δ2g x y = genotype x year variance; δ2e = environment variance; δ2p = phenotypic variance; MS g = mean square of genotypes; MS g x y = mean square of genotype x year; MS e = mean square of error; r = number of replications and y = number of year.
Both genotypic and phenotypic coefficients of variability were estimated using the formula below:
Phenotypic coefficient of variation 
Genotypic coefficient of variation 
δ2 p = phenotypic variance; δ2 g = genotypic variance;  = means of characters
= means of characters
Heritability in broad sense (h2) was computed as the ratio of genetic variance to total phenotypic variance
h2 = δ2g/ δ2px 100
The genetic advance (GA) and genetic advance as percentage of mean (GAM) were estimate using the formula given below:
GA = h2 k
GAM= GA /  x 100
x 100
Where h2 = heritability in broad sense; k= selection differential which equal to 2.06 at 5% intensity of selection;  “= general mean of the character.
“= general mean of the character.
Results and Discussions
Analysis of variance
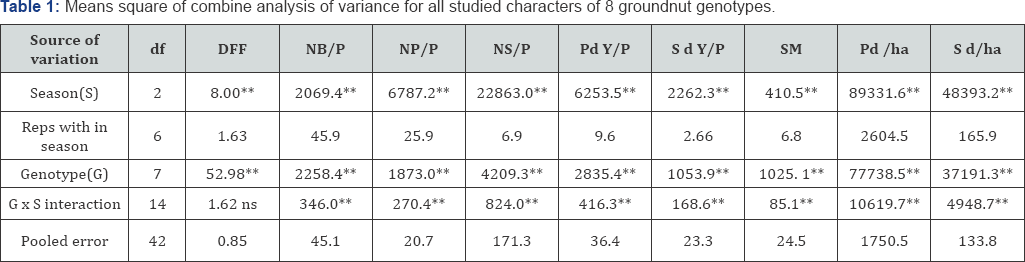
The results of combine analysis of variance for the all traits are shown in Table 1. High significant effects of genotypes were observed for all the characters under study indicating the existence of genotypic differences among the varieties. Highly significant variation was obtained due to season (year) for all the characters meaning that climate change were observed during the study. Environmental conditions were not similar during the investigation time. In addition to that, highly significant effect due to genotypes x season (year) was also obtained for all the characters except days to 50% flowering and plant height. This significant interaction effect of the traits under study indicated the genotypic differences in these characters were greatly influenced by the environment, on the other hand, non significant effect for interaction genotypes x season (year) exhibited for days to 50% flowering and plant height suggested less environmental influence on the expression of these traits. Similar results were obtained by Satish [20].
Range, mean, estimates of variance components and coefficients of variation
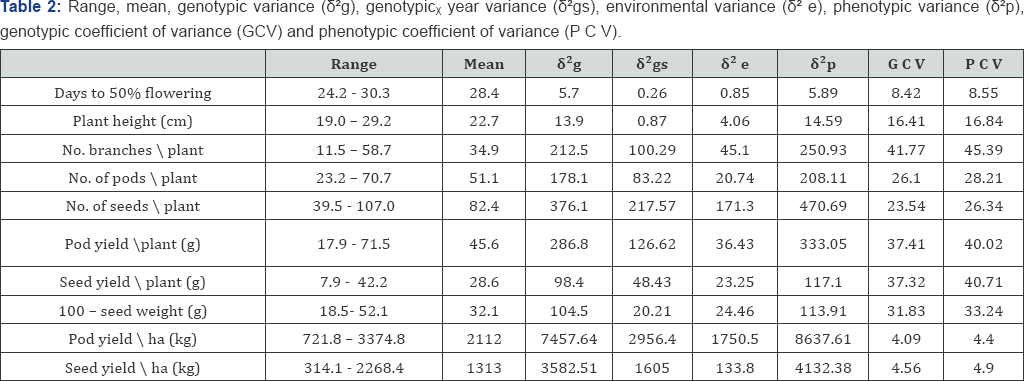
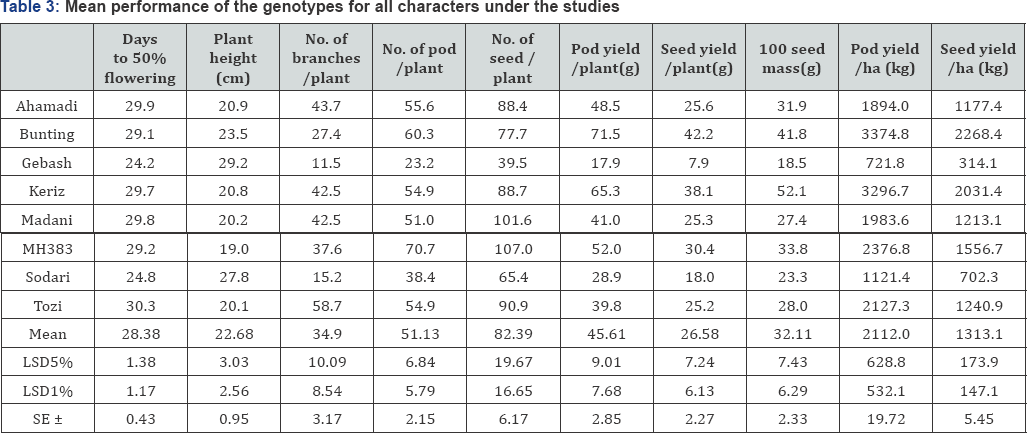
Range, mean, estimates of variance components and coefficients of variation, are shown in Table 2. All the characters exhibited wide range of variability except days to 50% flowering and plant height. Pod yield/ ha (kg) and seed yield/ ha (kg) which are the primary interest in most breeding program, showed a wide range of variation (3374.8-721.8) and (2268.4-314.1) respectively with mean value of (2112.0 and 1313.1) respectively In other side, the highest value of pod yield/ ha (kg) recorded by the genotype Bunting and the lowest value recorded by the genotype Gebash. Therefore, the presence of such range of variation of the traits indicated that the presence of large amount of genetic variation among the detected varieties which is the source of variable genetic material. In addition to that, all the characters exhibited larger genotypic variance than environmental variance indicated that the variation in phenotypic expression of the varieties is more due to genetic constituent and the contribution of the environment is fairly small.
The relative amount of variability in a population is best expressed in term of genotypic coefficient of variation, since this variable takes into account the mean values as well as the unit of measurement. The analysis of variation returned high estimates of genotypic coefficient of variation (GCV) for all characters studied except days to 50% flowering, pod yield/ ha, seed yield/ ha showed low GCV and plant height which exhibited moderated estimation for GCV. Similar trend were observed for phenotypic coefficient of variation (PCV). These are comparable to the results reported by Satish [4] & Gupta et al. [20]. Close relationship between GCV and PCV was found in days to % flowering, plant height, pod yield/ ha, seed yield/ ha; and PCV values were slightly greater than GCV revealing very little influence of environment for their expression similar results were reported by Satish [4], Gupta et al. [20] & Zaman et al. [21].
Heritability, genetic advance and genetic advance as percentage of mean
Heritability, genetic advance and genetic advance as percentage of mean are depicted in Table 3. Heritability plays a vital role in deciding the suitability and strategy for selection of a particular character. The traits under the study exhibited high broad sense heritability (96.94-84.01). Similar results reported by similar findings were reported by Gupta et al. [20] & Nath Uk, Alam [22].
Genetic advance (GA) under selection refers to the improvement of characters in genotypic value for the new population compared with the base population under one cycle of selection at given selection intensity. Highest value of GA was recorded with number of seed / plant (35.7) and the lowest (3.6) with plant height. The estimated value of excepted genetic advance expressed as percentage of mean ranged from (7.8- 72.9). Among the characters high values of GAM (>20%) were recorded for number of pod/ plant, number of seed/ plant, pod yield/ plant, seed yield/ plant and 100 seed mass, moderate value (10-20) % recorded by days to 50% flowering and plant height, and the low value of GAM (<10) obtained by pod yield/ ha and seed yield/ ha Similar results were obtained by Satish [4], Gupta et al. [20] Zaman et al. [21] & Nath Uk, Alam [22]
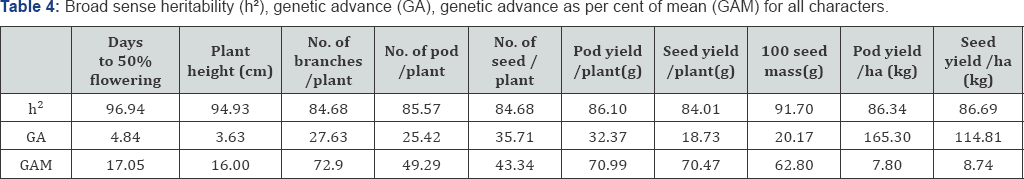
Reported that high values of genetic advance are indicative of additive gene action whereas low values are indicative of nonadditive gene action [23]. However, Jonson et al. [19] suggested that heritability estimates along with genetic advance would be more useful in predicting yield under phenotypic selection than heritability estimates alone. Except days to 50% flowering, plant height, pod yield/ha and seed yield/ ha, all the character showed high heritability estimates accompanied with high genetic advance. Therefore, direct selection based on phenotypic observations may be effective for improvement of these traits. Shannon [1] explains the magnitude of genetic variation among genotypes and the degree of heritability for the character to develop salt tolerant genotype(s).
Conclusion
The study revealed the existence of significant genetic variability among the tested varieties for different traits. The presence of significant genetic variability among the varieties suggested the possibility of improving the traits through direct and indirect selection. Traits with high heritability and genetic advance are important traits which should be given attention in order to bring an effective response of yield improvement of the concerned varieties under saline sodic soil.
Recommendations
a. The yield characters under investigation could successfully be selected among the genotypes for improvement in saline environment.
b. Genetic differences among genotypes give great opportunities to use these genotypes as a source of genetic material for the development of salt tolerance variants.
c. The study clearly demonstrated that there are a few high yielding genotypes that can endure satisfactory yield under salin sodic conditions; these genotypes could be find their way in to future groundnut breeding program.
References
- Shannon MC (1998) Adaptation of plants to salinity. Adv Agro 60: 7581.
- Munns R, (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25: 239-250.
- Abdallah HH (1986) Extent and management of problem soil in Sudan saline sodic soil, World Soil Resources Research 58: 90-109.
- Gupta RP, Vachhani JH, Kachhadia VH, Vaddoria MA, Papi Reddy (2015) Genetic variability and heritability studies in Virginia groundnut (Arachis hypogaea L.). Electronic Journal of Plant Breeding 6(1): 253256.
- Singh AI, Hariprassana k, Solanki R (2008) Screening and selection of groundnut genotypes for tolerance of soil salinity. Australian J of crop Sci 1(3): 69-77.
- Singh BG, Rao PV, Hiremath SM, (1989) Effect of salinity on germination and seedling growth in three varieties of groundnut (Arachis hypogaea L.). J Res APAU 17: 432-433.?
- Janila P, Rao TN, Kumar AA (1999) Germination and early seedling growth of groundnut (Arachis hypogaea L) varieties under salt stress. Annals Agric Res 20: 180-182.
- Senthil A, Djanaguiraman M, Vijayalakshmi C (2005) Influence of seed treatment of growth regulation on some enzyme activity in groundnut under salinity. Agricultura Tropica Et Subtropica 38(2): 88-89.
- Vadez V, Srivastava N, Krishnamurthy L, Aruna R, Nigam SN (2005) Standardization of a protocol to screen for salinity tolerance in groundnut. International Arachis News letter 25: 42-47.
- Singh R, Issar D, Zala PV, Nautigal PC, (2007) Variation in sensitivity to salinity in groundnut cultivars during seed germination and early seeding growth. ICRISAT 5(1): 1-7.
- Ahmed MSH, Mohamed SMS (2009) Improvement of groundnut (Arachis hypogaea L.) productivity under saline condition through mutation induction. World J of Agric Sci 5(6): 680-685.
- Foold MR, Jones RA (1993) Mapping salt-tolerance gene in tomato (Lycopersicon esculentum) using trait-based marker analysis. Theoretical Applied Genetics 87(1-2): 184-192.
- Ahmed MSH, Mohamed SMS (2009) Improvement of groundnut (Arachis hypogaea L) productivity under saline condition through mutation induction. World J of Agric Sci 5(6): 680-685.
- Winicov I, Bastola (1997) Salt tolerance in crop plants: new approaches through tissue culture and gene regulation. Acta physiologiae plantarum 1994): 435-449.
- Dudly JW, Moll RH, (1969) Interpretation and use of estimates of heritability and genetic variance in plant breeding. Crop Sci. 9: 257267.
- Falconer DS, Mackay FC, (1996) Introduction to Quantitative Genetics. Longman, New York, USA, pp. 464.
- Gomez KA, Gomez AA (1984) Statistical procedures for Agricultural Research. (2nd edn). Canada John Wiley and Sons Inc. 680.
- Burton GW, De Vane FH (1953) Estimation heritability in tall fescue (Fasculata arundinacea) from replicated clonal material. Agronomy Journal 45: 478-481.
- Johnson HW, Robinon HF, Comstock RE (1955) Estimation of genetic and environmental variability in soybean. Agron J 47: 314-318.
- Satish Y (2014) Genetic variability and characters association studies in Groundnut (Arashis hypogeae L). International Journal Of Plant, Animal And Environmental Sciences 4(4): 298-300.
- Zaman MA, Tuhina katun M, Ullah Z, Moniruzzamn M, Alam KH (2011) Genetic variability and path analysis of groundnut (Arachis hypogaea L). The Agriculturists 9(1-2): 29-36.
- Nath UK, Alam MS, (2002) Genetic Variability, Heritability and Genetic Advance of Yield Related Traits of Groundnut (Arachis hypogaea L). Online Journal of Biological Sciences 2(11): 762-764.
- Singh P, Marayanan SS (1993) Biometrical techniques in plant breeding. Kalyani publishers, New Delhi, India, p. 14-84.






























