Risk-Associated Circulating Biomarker ANXA2 Identified in Severe Acute Pancreatitis
Zhongsu Yu1, Huajian Hu1, Lu Xiao1, Xiaofeng Ma2 and Liangping Cheng1*
1Department of Gastroenterology Children’s Hospital of Chongqing Medical University, National Clinical Research Center for Child Health and Disorders, Ministry of Education Key Laboratory of Child Development and Disorders, China
2Department of Gastroenterology, Guangzhou Tianhe District People’s Hospital, Guangzhou, China
Submission:September 12, 2025;Published:December 09, 2025
*Corresponding author:Liangping Cheng, Department of Gastroenterology Children’s Hospital of Chongqing Medical University, National Clinical Research Center for Child Health and Disorders, Ministry of Education Key Laboratory of Child Development and Disorders, Chongqing 400000, China
How to cite this article:Zhongsu Yu, Huajian Hu, Lu Xiao, Xiaofeng M, Liangping C. Risk-Associated Circulating Biomarker ANXA2 Identified in Severe Acute Pancreatitis. Adv Res Gastroentero Hepatol, 2025; 22(2): 55607981.DOI: 10.19080/ARGH.2025.22.556081.
Abstract
Background: Specific single nucleotide polymorphisms (SNPs) can modulate gene expression and influence acute pancreatitis (AP) severity, so it’s crucial to understand AP’s genetic features.
Aims: This study aims to investigate the role of SNPs in the ANXA2 gene in severe acute pancreatitis (SAP), focusing on their associations with protein quantitative trait loci (pQTL) and their impact on gene expression.
Methods: A Summary-data-based Mendelian Randomization and publicly available Genome-Wide Association Study (GWAS) datasets were leveraged to identify SAP-related genetic variants. SNPs in the ANXA2 gene were selected based on their significant associations with pQTL in blood samples. These variants were pruned to ensure independence and reduce correlation, and a comprehensive analysis was performed on gene expression data from mouse and human samples.
Results: Our analysis found significant associations between specific ANXA2 SNPs and SAP severity. The identified SNPs could influence gene transcription and modulate circulating protein levels. Furthermore, a robust correlation was noted between immune cell infiltration and the expression of SAP-related genes, suggesting the potential role of immune responses in SAP progression.
Conclusion: The paper stresses that ANXA2 SNPs affect SAP severity through gene expression and immune responses. Future research should explore their mechanisms in SAP and inform therapies.
Keywords:Severe acute pancreatitis; ANXA2 gene; Single Nucleotide Polymorphisms; Mendelian Randomization; Protein quantitative trait loci; Immune response
Abbreviations:SNP: Single Nucleotide Polymorphisms; AP: Acute Pancreatitis; pQTL: Protein Quantitative Trait Loci; SAP: Severe Acute Pancreatitis; GWAS: Genome-Wide Association Study; MR: Mendelian Randomization; IV: Instrumental Variables; UKB-PPP: UK Biobank Pharma Proteomics Project; SMR: Summary-data-based Mendelian Randomization
Highlights:a) First identified: ANXA2 SNPs rs62004990 and rs8033800 affect transcription and protein levels.
b) ANXA2 highlight immune microenvironment’s role in SAP pathology.
c) ANXA2 identified as core regulatory factor in SAP progression.
d) Three downregulated miRNAs inhibit ANXA2 in patients.
e) Using two-sample Mendelian randomization to establish a robust model.
Introduction
Acute pancreatitis (AP) is a significant clinical condition that manifests as inflammation of the pancreas; with an estimated incidence of 34 per 100;000 individuals in the United States. It affects approximately 300;000 individuals each year and causes a case-fatality rate of 5% to 10% depending on the severity [1]. The pathogenesis of AP involves various genetic; environmental; and dietary factors. Genetic variations; particularly single nucleotide polymorphisms (SNPs); have been identified as critical contributors to the susceptibility to AP [2]. For example; studies have elicited that specific SNPs can modulate gene expression and influence AP severity [3]. Thus; understanding the genetic features of AP is crucial for developing targeted therapeutic strategies. Traditional statistical methods; such as regression analysis; have been widely used for studying the correlation between genetic variants and disease outcomes. However; these methods are limited due to confounding factors; reverse causation; and measurement errors [4]. For example; in observational studies; it is challenging to explain the effects of various environmental factors that may influence both exposure and the outcome. Hence; more reliable analytical methods are needed to determine causal relationships; better consider these confounding variables; and identify the genetic determinants of diseases like AP [5]. AP is a common disease of the gastrointestinal tract and the incidence is increasing at an annual rate of 2% to 5% [6]. Although the mortality rate has decreased over time; the overall mortality rate remains high. Severe AP (SAP) is strongly associated with mortality. Therefore; it is critical to predict risk factors for SAP.
Mendelian randomization (MR) is a potent statistical method that uses SNPs as instrumental variables (IVs) to infer causality between exposures and outcomes [7]. It utilizes the random assortment of alleles at conception; minimizes confounders; and estimates the causal effects more accurately [8]. Through genetic data; MR can uncover the impact of specific exposures; such as SNPs; on disease outcomes; thus, providing a better understanding of biological mechanisms. This approach is particularly important for studying complex diseases like AP; where traditional methods may fall short in establishing causality [1]. In this study; we aim to explore the causal link between genetic variants in the ANXA2 gene and the risk of developing SAP using a two-sample MR framework. The publicly available genome-wide association study (GWAS) data were leveraged to identify SNPs in the ANXA2 gene and assess their impact on AP severity [9]. We focus on how these SNPs modulate gene expression and influence disease outcomes and consider potential confounders such as age; sex; and lifestyle factors. Robust MR methods; including inverse-variance weighting and MR-Egger regression; were employed to ensure the reliability of our findings [1].
This study aims to elucidate the role of genetic variants in the ANXA2 gene in SAP. We integrate genetic data with clinical outcomes to identify novel genetic markers that help predict disease severity and guide therapeutic interventions. The result may enhance our understanding of the genetic features of AP and provide implications for personalized medicine; potentially improving clinical management strategies for patients at risk of SAP [2]. The UK Biobank Pharma Proteomics Project (UKB-PPP) is a collaborative initiative involving the UK Biobank (UKB) and 13 biopharmaceutical companies; which aims to characterize the plasma proteomic profiles of 54;219 UKB participants. This project has comprehensively mapped protein quantitative trait loci (pQTLs) for 2;923 proteins and identified 14;287 major genetic associations. Among them; 85% are newly discovered; including ancestry-specific pQTL mapping for non-European populations. The decode database offers large pQTL data to study the associations between plasma proteins and genetics. These data aid in identifying genetic variants that influence protein expression and revealing biological pathways related to diseases. In summary; eQTLs and pQTLs; particularly the data provided by UKB and decode; are vital tools for studying the genetic basis of gene expression and protein levels.
Methods
Study design and data sources
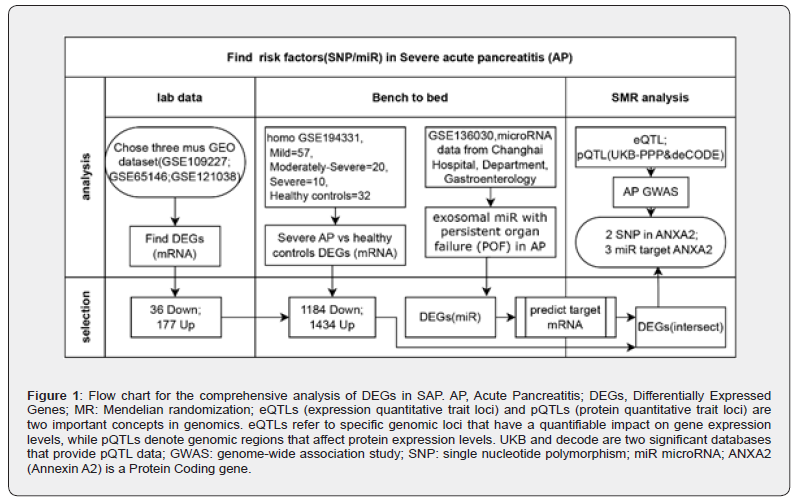
We applied a Summary-data-based Mendelian Randomization (SMR) combined Genome-Wide Association Study (GWAS) datasets and GEO datasheets data mining design to identify circulating proteins and regulated miRNAs associated with prevalence of SAP-related genetic variants (Figure 1). To achieve this; we used summary data from the largest genome-wide association study (GWAS) on AP conducted among individuals of European descent [1]; as well as protein quantitative trait loci (pQTL) GWASs from the Pharma Proteomics Project by Sun et al. [3] in the UK Biobank. Detailed methods of protein assays are described in this study [3].
Ethical approval
No separate ethical approval was required due to the use of publicly available data.
SNP selection and validation
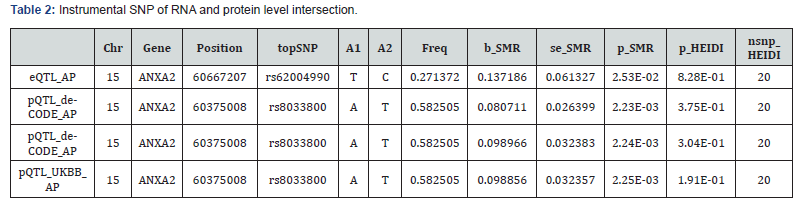
SNPs in the ANXA2 gene (GRCh38 chromosome 15 position 60;347;134-60;402;883 reverse strand) that was associated with gene expression (ie; pQTL) in blood samples were selected based on genome-wide significance (P<5×10−8). To ensure minimal linkage among the IVs used in MR; they were ranked based on the P value of their associations with UKB_PPP_pQTL (https:// registry.opendata.aws/ukbppp/)[10] and subsequently pruned for linkage disequilibrium at an r2 threshold of 0.1 and a distance threshold of 500 kilobases. Considering the correlation between variants; including mildly correlated variants; can improve statistical power over strictly selecting independent variants. MR estimators become unstable when only selecting variants that are highly correlated [11]. The pruning r2 threshold was used to balance these two issues.
MR analysis
The MR approach followed three key assumptions [12]. First; the IVs used were highly correlated with exposure. Largescale modern GWASs can identify genetic variants associated with exposure in large datasets [13]. Second; the IVs must not be affected by any confounders. This assumption may be violated due to LD and/or population ancestry [14]. Lastly; IVs must only affect the outcome through exposure (namely; no horizontal pleiotropy) [15]. Large-scale GWASs of circulating proteins [16] have evidenced that their genetic determinants are near the encoding genes. The cis-acting SNPs reduce potential horizontal pleiotropy and enhance the validity of MR assumptions. A cis-SNP that is closely related to the protein is likely to directly affect gene transcription and circulating protein levels. Independent cis-pQTL SNPs (r2≤0.001) that were notably correlated with circulating proteins (p<5×10−8) were selected from pQTL GWASs [16].
Statistical Analysis
Data were analyzed in R software 4.3.3. Continuous data were compared using independent Student’s t-test or Mann-Whitney U. Differences among groups were compared via the Kruskal- Wallis test. Spearman correlation analysis was utilized to obtain coefficients among genes. A p-value (two-sided) < 0.05 implied statistical significance.
Software and Preregistration
MR analyses were conducted using the SMR & HEIDI methods and software tool [11,16]. Some data from GWAS were extracted from the OpenGWAS platform. This paper was not pre-registered.
Results
Data acquisition
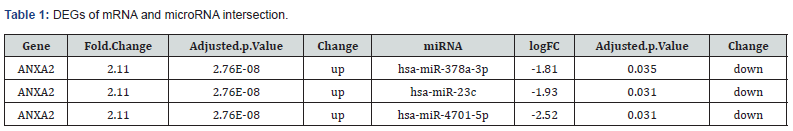
Data from the GEO database were downloaded using the R package GEOquery [17]. Five AP datasets (GSE109227; GSE65146; GSE121038; GSE136030; and GSE194331) were obtained. The samples from the GSE109227 [18]; GSE65146; and GSE121038 datasets were all derived from the pancreas tissue of mice. The samples from the GSE136030 (GEO Accession viewer) and GSE194331 [19] datasets were all derived from human exosomal miRNA and RNA-Seq of blood collected from AP patients with varying severity. The detailed information is listed in Figure 1. In the GSE109227 dataset; the chip platform was GPL6246; which included 6 AP samples and 5 control samples. In the GSE65146 dataset; the chip platform was also GPL6246; including 39 AP samples; 5 control samples; and 27 mutant mouse samples. The GSE121038 dataset utilized the GPL10787 chip platform and included 8 AP samples and 7 control samples. This study only considered the AP group and the control group. In the GSE136030 dataset; the chip platform was GPL18402. Exosomal miRNAs in serum samples from AP patients with or without persistent organ failure (POF) (5 vs. 5) were profiled using microarrays and differentially expressed genes (DEGs) were identified. Then starBase v2.0 [20] was utilized to get the target mRNA. The GSE194331 dataset [19] utilized the GPL16791 chip platform and included 87 AP patients of varying severity levels (Mild=57; Moderately-Severe=20; Severe=10) and 32 healthy controls. Deseq2 was utilized to get DEGs between SAPs and healthy controls (Table 1).
The sva [21] R package 3.50.0 was used to eliminate batch effects from the GSE109227; GSE65146; and GSE121038 datasets; resulting in a combined dataset; which encompassed 53 AP cases and 17 control cases. Subsequently; the combined dataset was standardized with the R package limma [22]. Probes for descriptions were optimized and uniformly produced. Principal component analysis (PCA) was then carried out on expression matrices to verify the effectiveness of the elimination process. PCA reduced data dimensionality; involved feature vectors (components) from high-dimensional data; and represented them in a lower-dimensional space in two-dimensional or threedimensional plots. The AnnoProbe 0.1.7 (https://github.com/ xjsun1221/tinyarray) R package were used to analyze exosomal miRNAs; which were quantified in each group for AP individuals with POF at an early phase. Deseq2 (https://github.com/ thelovelab/DESeq2) was used to analyze the GSE194331 dataset; and transcriptomic data found increased DEGs correlated with mild; moderate; and SAP; demonstrating 447; 1;553; and 2;618 DEGs; respectively (padj≤0.05; logFC ≥2).
DEGs Related to SAP
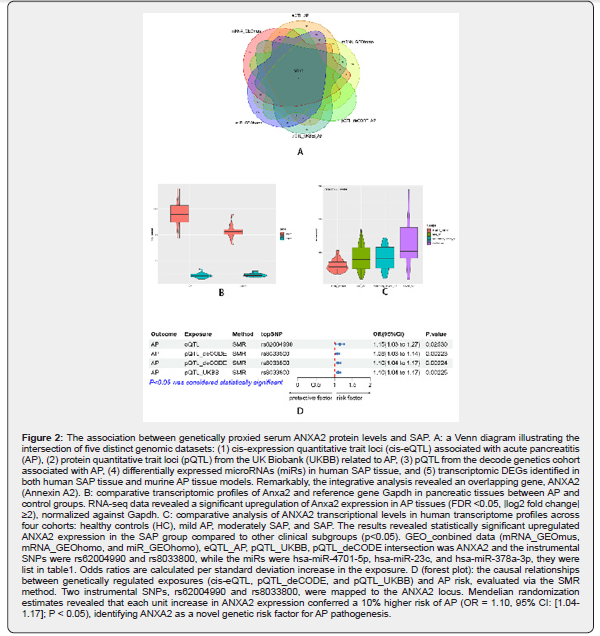
Based on samples from the GEO database; mouse samples were classified into the control and AP groups. DEGs between the two groups were analyzed via the R package; with |logFC|>1 and adj.p<0.05 as the criteria. Genes with logFC>1 and adj.p<0.05 were considered up-regulated; while genes with logFC<-1 and adj.p<0.05 were classified as down-regulated. The Benjamini- Hochberg (BH) approach was used to obtain adj.p value. Volcano plots were employed to display DEGs. DEGs in human samples were identified as genes with |logFC| ≥2 and adj.p ≤ 0.05 (corrected using the BH method). To locate SAP-related genes; SMR SNP instrument intersection genes were overlapped with DEGs from the combined dataset. A Venn diagram was constructed to demonstrate the overlapping genes (Figure 2A). These genes were further visualized in a boxplot from mouse data (Figure 2B) and human data (Figure 2C). Forest plots showed the top SNP from eQTL and pQTL was rs62004990 and rs8033800; increasing the risk of SAP to 10% (Figure 2D).
Immune Infiltration Analysis
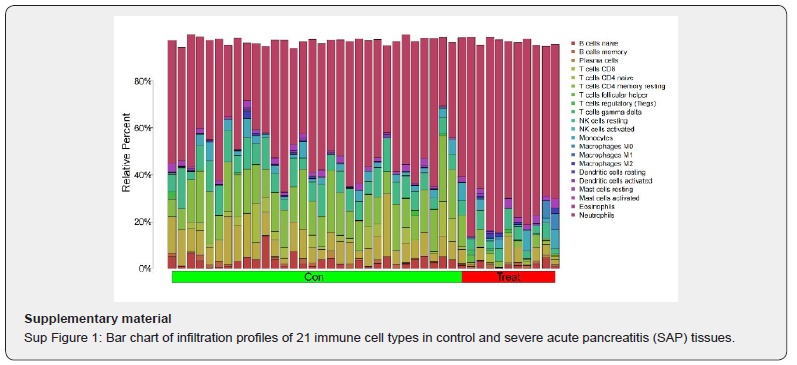
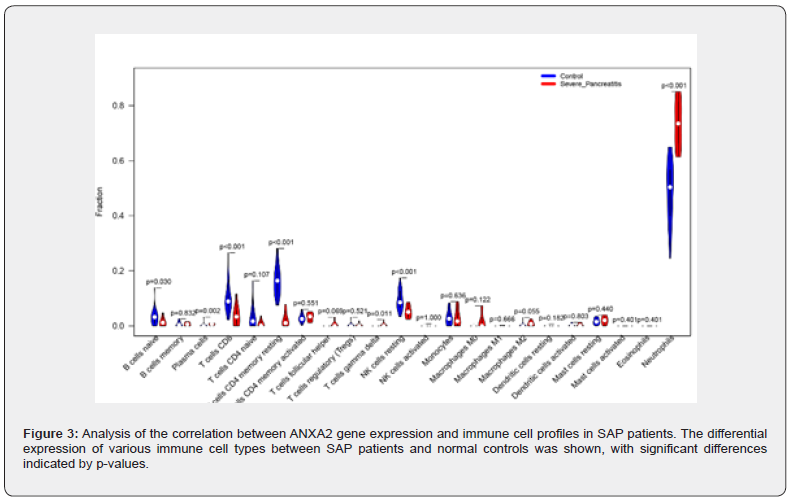
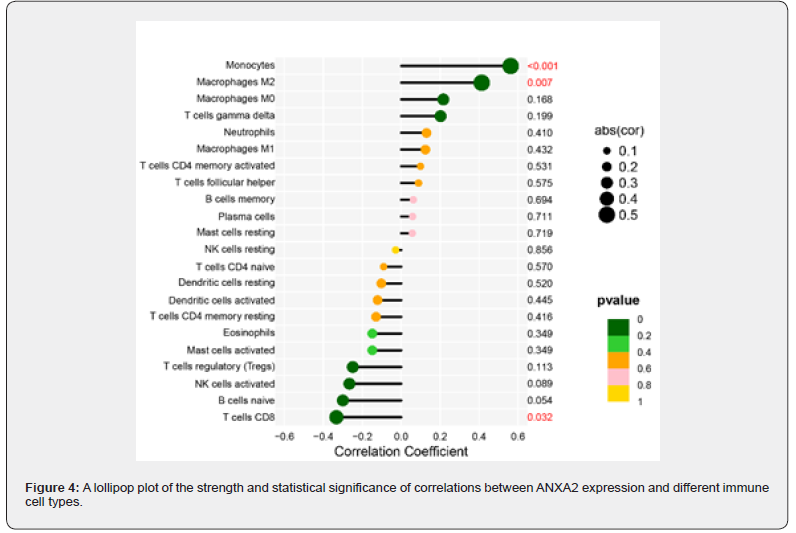
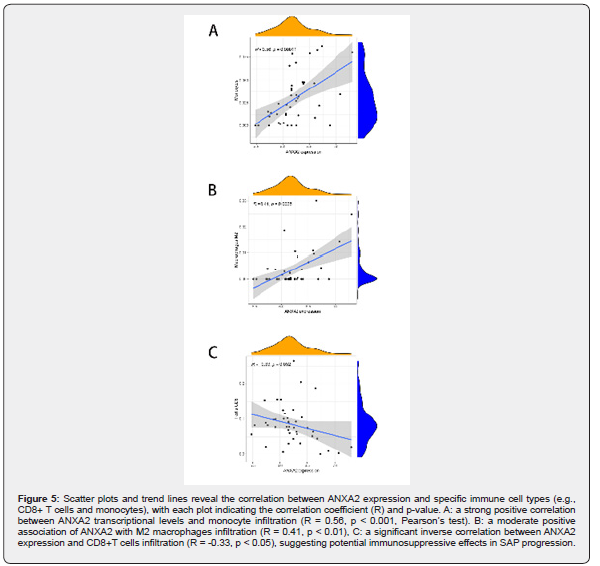
Based on linear support vector regression; the CIBERSORT algorithm [23] deconvolutes the transcriptome expression matrix to estimate the composition and abundance of immune cells in cell mixtures. Data with an immune cell enrichment score > zero were filtered and the specific immune cell infiltration in the matrix of the combined dataset was obtained. The proportion of immune cells was visualized in a bar chart (sup Figure 1 in supplementary material). The different immune cells between healthy controls and SAP patients were shown in plots using the R package ggplot2. Subsequently; the relevance was obtained by the Spearman algorithm and visualized in a correlation scatter plot. Additionally; the Spearman algorithm was utilized to obtain the relevance of immune cells to the focused gene; which was visualized in the violin plot (Figure 3); lollipop plot; and scatter plot (Figure 4 & Figure 5). Our study found that the overlapping gene identified from the combined analysis of GEO data; eQTL_AP; pQTL_UKBB; and pQTL_deCODE was ANXA2. The three miRNAs downregulated in extracellular vesicles from the peripheral blood of SAP patients could target ANXA2. These miRNAs were hsamiR- 378a-3p; hsa-miR-23c; and hsa-miR-4701-5p; with hsa-miR- 4701-5p showing the most significant decrease; with a logFC of -2.52 and an adjusted p-value of 0.035 [24] (Table 2).

Discussion
This study investigated the genetic basis of AP and its impact on AP severity. Our findings revealed a significant association between specific genetic variants and AP severity; as evidenced by identified DEGs across various datasets. Notably; the analysis highlighted the role of immune cell infiltration in the pathophysiology of SAP; suggesting that genetic predispositions may influence immune responses during SAP progression. These results stress the importance of the genetic features of AP; which could pave the way for targeted strategies and improved management. Our findings align with previous research that underscores the role of cis-acting SNPs in the risk of AP. For instance; Wang et al. [25]. employed the MR approach to investigate the causal link between lipid levels and AP and revealed that higher triglyceride levels significantly increased the risk of AP (Odds Ratio [OR] = 2.02; 95% CI: 1.22-3.31)[25]. Additionally; Kim et al. identified genetic variants that influenced immune traits associated with AP; suggesting that immune dysregulation may be crucial in AP pathogenesis [26]. Similarly; Zhang et al. found several significant genetic loci associated with the susceptibility to AP; providing strong evidence for the genetic basis of AP [27] (Table 3).
Furthermore; recent findings indicate a complex interaction between genetic factors and environmental influences in determining AP risk. For instance; Chuang et al. explored the association between gut microbiota and AP; revealing that specific microbiota profiles; driven by genetic predisposition; could impact the risk of AP [28]. This illustrates that both genetic factors and environmental interactions contribute to AP risk; consistent with our emphasis on the multifactorial nature of AP. In contrast; other studies report the opposite findings. For example; Li et al. observed an association between specific SNPs and an increased risk of AP; yet they found no causal link through MR analyses [29]. This discrepancy might be attributable to differences in methodology; sample size; or population diversity. Similarly; Zhang et al. indicated that while dietary factors appeared to influence AP risk; they did not find significant genetic associations [30]. These inconsistencies underline the intricate nature of genetic interactions in AP and further investigations are required to clarify these relationships. Moreover; Wang et al. supported a significant genetic predisposition to AP-related immune traits; consistent with our findings [11]. Conversely; the lack of found by Liu et al. [26]. did not reveal a genetic association in their study of obesity-related factors. Hence; more research is required to fully illustrate the interplay between genetic and environmental factors in AP pathogenesis [31].
In summary; our findings corroborate existing literature on the genetic factors for the risk of AP; particularly emphasizing the role of cis-acting SNPs and immune-related genetic traits. The inconsistencies observed should be explained in future investigations; particularly concerning the methods employed and the populations studied; to enrich our comprehension of the genetic architecture of AP. The association between ANXA2 SNPs and AP is elucidated through its influence on gene transcription and subsequent circulating protein levels. The ANXA2 gene; which encodes annexin A2; is crucial in various cellular processes; including membrane trafficking and inflammation; both of which are pivotal in AP pathogenesis. Studies have noted that SNPs in the ANXA2 gene can affect its transcriptional activity; thereby potentially altering the expression levels of annexin A2 under APrelated inflammatory stimuli [32]. Notably; alterations in annexin A2 levels can regulate pancreatic inflammation and fibrosis; suggesting a direct link between ANXA2 expression and AP severity [7]. Furthermore; the interaction of ANXA2 with various signaling pathways underscores its significance in inflammatory responses. For instance; ANXA2 activates pathways involved in the release of pro-inflammatory cytokines; thereby influencing the acute-phase response in AP [33]. ANXA2 also promotes cell survival during inflammatory processes; showing potentially protective mechanisms against pancreatic acinar cell damage; which is a hallmark of AP [34].
ANXA2 dysregulation through genetic variants may impair cellular responses and ultimately contribute to the pathological mechanisms of AP. Specifically; increased levels of ANXA2 can promote the recruitment of immune cells to the inflammatory site; thus, exacerbating local inflammatory responses [35]. Additionally; circulating ANXA2 levels may be a biomarker for AP severity; providing clinical implications for detection and therapeutic interventions [36]. In conclusion; the SNPs in the ANXA2 gene significantly influence the transcription and subsequent protein levels; which in turn modulate the pathological mechanisms of AP. The intricate relationship between ANXA2 expression and inflammatory processes warrants further investigations to determine its potential as a therapeutic target and biomarker in AP management. Future studies should explore the precise mechanisms of ANXA2 SNPs in gene transcription and clinical implications for AP patients. Understanding these pathways could offer new ideas for therapeutic strategies to mitigate the inflammatory response in AP [37].
This study employs MR analysis and large-scale GWAS data; presenting a significant advantage over previous research that often lacked rigorous methodologies to address confounding factors. The two-sample MR approach effectively reduces the influence of confounders and enhances the reliability and validity of our results. Unlike earlier studies that primarily relied on observational data without genetic profiles; our approach more accurately estimates the causal relationship. Multiple MR methods; such as inverse variance weighting and sensitivity analyses are used; which further confirms the robustness of our findings and ensures that the results are not influenced by pleiotropy or heterogeneity. Previous studies have demonstrated challenges in establishing causality due to confounding factors. However; we obtain consistent findings across various analytical techniques; reinforcing the strength of our conclusions. Additionally; specific genetic variants were regarded as IVs; which enhances the accuracy of our causal inferences and elucidates the underlying mechanisms. Overall; this research offers a reliable framework for exploring causality and valuable insights into the complex impact of genetic factors on health outcomes [16,38,39].
This study highlights the significant association between specific DEGs and AP severity; suggesting a potential role of these DEGs in the AP pathophysiology. Despite these important findings; several limitations must be acknowledged. First; the absence of wet lab experiments restricts the biological validation of the identified DEGs. Additionally; the relatively small sample sizes in some datasets may limit the generalizability of our conclusions. Given the lack of clinical validation; greater caution must be exercised in interpreting the results. Furthermore; integrating multiple datasets may introduce batch effects that affect the consistency of data analysis.
In conclusion; our research confirmed the association between immune infiltration and specific gene expression patterns in SAP; underscoring the importance of these factors for future therapeutic strategies. These findings should be substantiated in clinical settings to disclose the underlying mechanisms of AP. Larger-scale studies and clinical validation are essential to confirm the relevance of these DEGs as biomarkers and potential therapeutic targets.
References
- Yang Y, Hu P, Zhang Q (2024) Single-cell and genome-wide Mendelian randomization identifies causative genes for gout. Arthritis Res Ther 26(1): 114.
- Bourgault J, Abner E, Manikpurage HD (2023) Proteome-Wide Mendelian Randomization Identifies Causal Links Between Blood Proteins and Acute Pancreatitis. Gastroenterology 164(6): 953-965.
- Zhao T, Guan X, Hu Y (2024) Population-wide DNA methylation polymorphisms at single-nucleotide resolution in 207 cotton accessions reveal epigenomic contributions to complex traits. Cell Res 34(12): 859-872.
- Chen J, Chen H, Mai H (2023) A functional variant of CD40 modulates clearance of hepatitis B virus in hepatocytes via regulation of the ANXA2/CD40/BST2 axis. Hum Mol Genet 32(8): 1334-1347.
- Hartanto M, Sami AA, De Ridder D, Nijveen H (2022) Prioritizing candidate eQTL causal genes in Arabidopsis using Random Forests. G3 (Bethesda) 2(11).
- Balint ER, Fur G, Kiss L (2020) Assessment of the course of acute pancreatitis in the light of aetiology: a systematic review and meta-analysis. Sci Rep 10(1): 17936.
- Yi H, Yang Q, Repaci C (2024) TOPMED imputed genomics enhances genomic atlas of the human proteome in brain, cerebrospinal fluid, and plasma. Sci Data 11(1): 387.
- Abbas M, Goodney G, Vargas JD, Gaye A (2024) Transcriptome Study of 2 Black Cohorts Reveals cis Long Noncoding RNAs Associated With Hypertension-Related mRNAs. J Am Heart Assoc 13(11): e034417.
- Niu HM, Yang P, Chen HH (2019) Comprehensive functional annotation of susceptibility SNPs prioritized 10 genes for schizophrenia. Transl Psychiatry 9(1): 56.
- Sun BB, Chiou J, Traylor M (2023) Plasma proteomic associations with genetics and health in the UK Biobank. Nature 622(7982): 329-338.
- Woolf B, Raja Sundaram S, Cronje HT, Yarm Olinsky J, Burgess S (2023) A drug target for erectile dysfunction to help improve fertility, sexual activity, and wellbeing: mendelian randomization study. BMJ 383: e076197.
- Strobe-MR: guidelines for strengthening the reporting of mendelian randomization studies.
- Tam V, Patel N, Turcotte M, Bosse Y, Pare G (2019) Benefits and limitations of genome-wide association studies. Nat Rev Genet 20(8): 467-484.
- Swanson SA, Hernan MA (2018) The challenging interpretation of instrumental variable estimates under monotonicity. Int J Epidemiol 47(4): 1289-1297.
- Davies NM, Holmes MV, Davey Smith G (2018) Reading Mendelian randomization studies: a guide, glossary, and checklist for clinicians. BMJ 362: k601.
- Zhu Z, Zhang F, Hu H (2016) Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 48(5): 481-487.
- Davis S, Meltzer PS (2007) Geoquery: a bridge between the Gene Expression Omnibus (GEO) and Bioconductor. Bioinformatics 23(14): 1846-1847.
- Norberg KJ, Nania S, Li X (2018) RCAN1 is a marker of oxidative stress, induced in acute pancreatitis. Pancreatology 18(7): 734-741.
- Nesvaderani M, Dhillon BK, Chew T (2022) Gene Expression Profiling: Identification of Novel Pathways and Potential Biomarkers in Severe Acute Pancreatitis. J Am Coll Surg 234(5): 803-815.
- Li JH, Liu S, Zhou H, Qu LH, Yang JH (2014) Starbase V2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42: D92-D97.
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD (2012) The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28(6): 882-883.
- Ritchie ME, Phipson B, Wu D (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7): e47.
- Newman AM, Liu CL, Green MR (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12(5): 453-457.
- Rhodes CJ, Batai K, Bleda M (2019) Genetic determinants of risk in pulmonary arterial hypertension: international genome-wide association studies and meta-analysis. Lancet Respir Med 7(3): 227-238.
- Wang B, Dron JS, Wang Y (2024) Lipid levels and risk of acute pancreatitis using bidirectional Mendelian randomization. Sci Rep 14(1): 6267.
- Liu R, Wang K, Guo X (2024) A causal relationship between distinct immune features and acute or chronic pancreatitis: results from a mendelian randomization analysis. Pancreatology 24(8): 1219-1228.
- Zhou F, Liu Y, Shi Y, Wu N, Xie Y (2024) Association between gut microbiota and acute pancreatitis: a bidirectional Mendelian randomization study. J Gastroenterol Hepatol 39(9): 1895-1902.
- Fu X, Wu H, Shu Y, Yang B, Deng C (2024) Crohn disease but not ulcerative colitis increases the risk of acute pancreatitis: A 2-sample Mendelian randomization study. Medicine (Baltimore) 103(23): e38317.
- Paquette M, Guay SP, Baass A (2024) Genetic determinants of pancreatitis risk in hypertriglyceridemia. Curr Opin Lipidol.
- Mao X, Huang C, Wang Y (2023) Association between Dietary Habits and Pancreatitis among Individuals of European Ancestry: A Two-Sample Mendelian Randomization Study. Nutrients 15(5).
- Rao M, Ai X, Huang Z (2023) The Causal Effects of Cholelithiasis on Acute Pancreatitis and Pancreatic Cancer: A Large Sample Size Mendelian Randomization Analysis. Recent Pat Anticancer Drug Discov.
- Baharom F, Ramirez-Valdez RA, Khalilnezhad A (2022) Systemic vaccination induces CD8(+) T cells and remodels the tumor microenvironment. Cell 185(23): 4317-4332.
- Fingerhut JM, Lannes R, Whitfield TW, Thiru P, Yamashita YM (2024) Co-transcriptional splicing facilitates transcription of gigantic genes. PLoS Genet 20(6): e1011241.
- Pathogenesis of acute pancreatitis. Der Internist.
- Mihoc T, Latcu SC, Secasan CC (2024) Pancreatic Morphology, Immunology, and the Pathogenesis of Acute Pancreatitis. Biomedicines 12(11).
- Bai R, Lu TQ, Sun B. Evolution and progress of surgical intervention strategies for acute pancreatitis. Zhonghua Wai Ke Za Zhi 2023;61(7):556-561.
- Petrov MS (2020) Panorama of mediators in post pancreatitis diabetes mellitus. Curr Opin Gastroenterol 36(5): 443-451.
- Wan X, Yu H, Yang M (2024) Study on the causal relationship between educational attainment and delirium: A two-sample Mendelian randomization study. Heliyon 10(7): e28697.
- He H, Liao S, Zeng Y, Liang L, Chen J (2022) Causal relationships between metabolic-associated fatty liver disease and iron status: Two-sample Mendelian randomization. Liver Int 42(12): 2759-2768.






























