Mini Review of Mice and Men: From Reductionism towards a Systems Biology Approach to Elucidate the Molecular Mechanism behind the Pathogenesis of Type 2 Diabetes
Vincent van Ginneken1,2*, Elwin Verheij3 and Clemens Lowik2
1Bluegreentechnologies, Heelsum, Netherlands
2Erasmus Medical Centre, Radiology and Nuclear Medicine, Optical Molecular Imaging, Netherlands
3TNO, Pharma, Zeist, Netherlands
Submission: February 03, 2018; Published: March 09, 2018
*Corresponding author: Vincent van Ginneken, Bluegreentechnologies, Heelsum, Erasmus Medical Centre: Radiology and Nuclear Medicine, Optical Molecular Imaging, Netherlands, Email: vvanginneken@hotmail.com
How to cite this article: Vincent van G, Elwin V, Clemens L. Mini Review of Mice and Men: From Reductionism towards a Systems Biology Approach to Elucidate the Molecular Mechanism behind the Pathogenesis of Type 2 Diabetes. Adv Res Gastroentero Hepatol 2018; 9(1): 555755. DOI: 10.19080/ ARGH.2018.09.555755.
Abstract
In this mini-review via a Reductionistic one gene approach with rodent models versus the System Biological approach with Lipidomics based LCMS techniques we will compare the molecular mechanisms of Insulin Resistance (IR) and Type 2 Diabetes (DM2). Through a reductionist one gene knock-in and knock-out approach with mouse models -starting from the eighties previous century- researchers succeeded to unravel the molecular mechanism of IR/DM2. Recently, via a System Biological LCMS lipidomics approach, our research group succeeded to find two clear biomarkers for IR/T2DM: a 20:3 Cholesterylester (dihomo-y-linolenic; 20:3 n-6) earlier classified as a DM2 biomarker in a human cohort of the Uppsala Longitudinal Study of Adult Men (ULSAM). The second biomarker was C36:1-Phosphatidylcholine for which we hypothesized its concentration rose significantly in non-adipose tissue due to the evolving hypoxic conditions as a consequence of a High-fat diet induced IR/ T2DM. As an earlier defined prerequisite, both compounds were not found in the food. This LCMS Systems Biology Lipidomics approach has large potential to find biomarkers for Liver cancer (HCC) such as primary liver cancer (PLC) organoids and its PLC subtypes: hepatocellular carcinoma (HCC), cholangiocarcinoma (CC) and combined HCC/CC (CHC) tumors. In future studies we will follow this approach using a novel, near-physiological organoid culture system with primary human healthy liver cells that retain liver tissue function and genetic stability.
Keywords: Type 2 diabetes; Insulin resistance; Mouse models; Reductionism; Systems Biology; biomarker; 20:3 Cholesterylester; C36:1 Phosphatidylcholine; Hypoxia; Hepatocellular Carcinoma (HCC); Organoid culture system
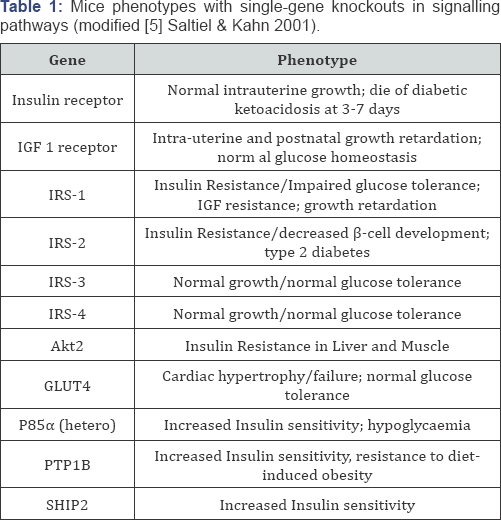
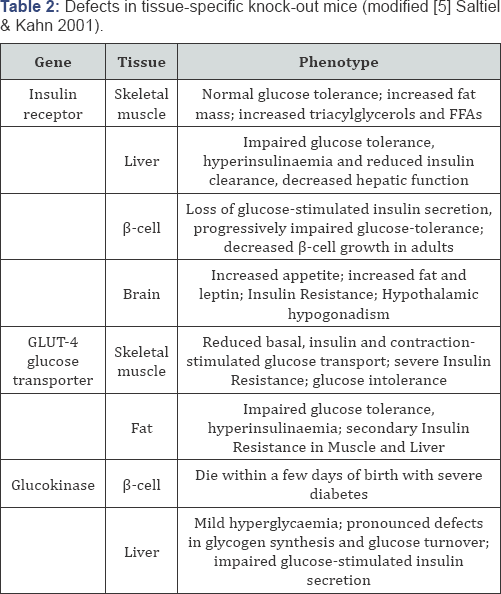
Introduction
Mainly via a reductionistic approach using animal mouse models of mainly genetically engineered mice, new insight with (single genes or cluster of genes) were obtained in the pathogenesis of Insulin Resistance (IR) and Type 2 Diabetes (DM2) [1]. By eliminating certain genes (knock-out) or inserting mutations into specific genes, creating transgenic animals, (knock-in) some genetic mechanisms became clear. The different mouse strains gave for single gene defects valuable information e.g. for syndromes of severe IR [2], mitochondrial diabetes [3], the different obese mouse strains like ob/ob, db/db and fa/ fa [1]. These mouse models have been added a very important contribution in understanding the different processes/ steps in the pathogenesis of DM2 [1] and Islet of Langerhans Degeneration [4]. In the review of Saltiel & Kahn [5] in Table 1 one a list of mice-phenotypes with single-gene knockouts in signalling pathways and the regulation of glucose and lipid metabolism is given regarding the gene for: the Insulin receptor, IGF-1 receptor, IRS-1, IRS-2, IRS-3 and IRS-4, Akt2, GLUT4, P85a (hetero), PTP1B and SHIP2. While in the same review in Table 2 the defects in tissue-specific knockout mice are given for the gene regarding: the Insulin receptor, GLUT4 and glucokinase [5]. These knockout mice models let us to the Insulin- stimulated glycogen synthesis and GLUT4 translocation: PDK/ PKB hypothesis [6,7]. This hypothesis proposes that binding of insulin to its receptor phosphorylates insulin receptor substrate 1 (IRS-1) causing it to attract p85, which binds to and activates phosphatidyllinositol-3 kinase (PI-3 kinase), which binds to and activates phosphatdidyllinositol-3 kinase (PI-3 kinase). This forms phosphatidyllinositol 3,4,5-trisphosphate, which binds both protein kinase B (PKB) and phosphoinositide-dependent kinase 1 (PDK-1), juxtapositioning them in the membrane and enabling PDK-1 to phosphorylate and activate PKB (=Akt). PKB in turn phosphorylates and inactivates glycogen synthase kinase 3 (GSK-3). Because GSK-3 is constitutively active and inhibits glycogen synthase, inactivation of GSK-3 permits glycogen synthesis. PKB alsophosphorylates proteins involved in GLUT4 translocation [6,7].
Coming back to the pathogenesis of DM2 which can be generally described by two main characteristics:
A) . Impaired ability of the pancreatic β-cells to produce or secrete insulin or even loss of β-cells [8] gave in his review 4 possible reasons: A-1). Fatty acids are converted in β-cells to toxic compounds like ceramides leading to p-cell loss/death; A-2). The fatty acid toxicity results in a failure of the β-cells to secrete insulin; A-3). Fatty acid toxicity can lead to an inactivation of the insulin receptor in the β-cells; A-4). Uncoupling protein-2 is a negative regulator of Insulin secretion.
B) . Diminished ability of insulin to exert its effect on the target tissue (muscle, adipose tissue and liver function). For the latter cause the following 4 major reasons can be mentioned [6,7]: B-1). The secretion of insulin of β-cells is inadequate (insufficient or too slow); B-2). The released insulin is structurally abnormal and thus only partial functional; B-3). Defects could be in the tissues targeted by Insulin especially skeletal muscle, adipose tissue and the liver; B-4). The insulin receptor is defective or the transduction and/or amplification of the insulin signal to intracellular metabolic processes is impaired.
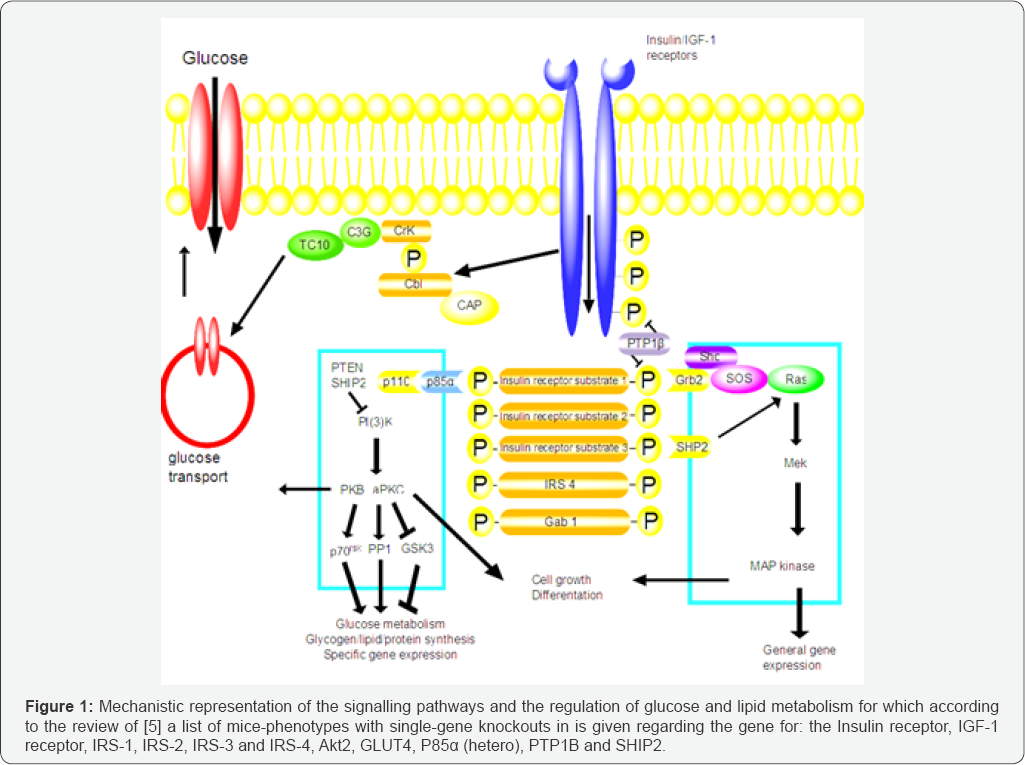
The reductionistic approach has led using animal rodent- models [1,4] has uncovered many potential candidate for diabetes susceptibility genes. However an "one single gene” approach might only in a few percentages of the DM2 be the cause. However, in general it as assumed DM2 is a polygenic disorder [9]. Therefore a Systems-Biology approach is clearly wARGHanted. The Reductionistic versus Systems-Biology approach is related to a discipline e.g. reductionism is propagated in Sciences like Chemical Engineering [10], Biochemistry and Biophysics [11], while Systems-Biology is propagated in Sciences like e.g. Biology and Medical-Biology [12]. The major "drive” and interest for a Systems-Biology approach is linked to the progress in collecting tremendous experimental datasets and at the system-level understanding of native biological and pathological systems to provide potential therapeutic targets [13,14] (Figure 1).
Medicine is at the edge, clinical medicine focuses on the parts and the Systems-Biology on the systems. While the "traditional” approach in medicine is based on four principles: 1). Focus on a singular, dominant factor which is the deeply rooted belief that each disease has a potential singular target for medical treatment. For example, an infection has to be treated with antibiotics but the underlying mechanism e.g. an impaired immune system is not treated. 2). Emphasis on homeostasis. This is based on two principles: 2a). Homeostasis needs to be maintained by placing a deviating physiological parameter (e.g. high blood pressure) within its physiological range; 2b). Reductionism often disregards the dynamic interaction between parts, the system is often depicted as a collection of static components. 3). Inexact risk modification: the often in medical epidemiology "one risk-factor to one-disease” approach has certain limitations. 4). Additive treatments which can be characterized by a reductionistic approach, an one risk to one-disease analysis which ultimately results the inability e.g. for diseases with multiple risk factors like Obesity or 1R/DM2 to calculate their collective influences [14]. The need to make sense of complex genetic interactions has led some researchers from a component-level to system-level perspective. In understanding the difficulty of complex biological systems research like e.g. a complex disease like DM2 has to be directed to the components of a biological system, their interactions and the behaviors and properties of the whole system. This is the basis for a Systems Biology approach which aims to understand phenotypic variation to assemble comprehensive data and models of cellular organization and biochemical function, and to elucidate interactions and pathways for e.g. metabolites [15]. The progress made in this new research area of Systems-Biology is also related to the progress made the last decade in other research area's like: Molecular Biology, Computational Science, Statistics, Chemistry, and Mathematics ([16] Figure 2).
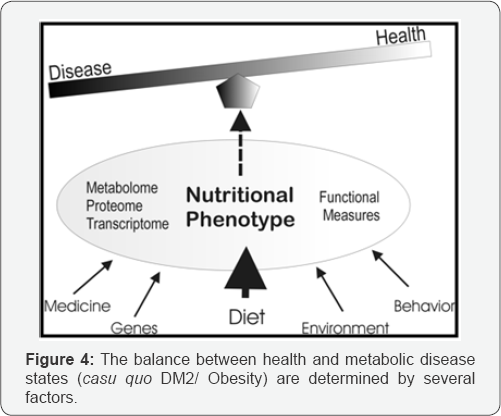
Metabolomics is a discipline dedicated to the systematic study of small molecules (i.e. metabolites) in cells, tissues, and different bio-fluids. Metabolite levels can be regarded as amplified responses of biological systems to genetic or environmental changes [17]. The term-omics represents the rigorous study of various collections of molecules, biological processes, or physiological functions and structures as systems [17]. The challenge of a “omics” approach is not only the discovery of changes in metabolite profile but it is figuring out what these changes mean which ultimately can lead to a biomarker for a pathogenesis (Figure 3). From Figure 4 we can see that the balance between “disease” or “healthy” is determined by factors like on one hand: environment, behavior and diet and for a diseased person by genetics and medical treatment.
In principle metabolomics can provide certain advantages relative to other -omics technologies (genomics, transcriptomics, proteomics) like is reviewed for DM2 research by Bain et al. [18]. A particular area of interest of Metabolomics based Systems- Biology, will be identification of novel safety biomarkers that can be used in the assessment of new intensive treatments both during their discovery phase and in clinical use to study e.g. metabolism-related diseases like 1R/DM2 in combining the diagnostics with the patient therapy [19]. Recent technological advances in metabolomics [20,21] and lipidomics [22] can potentially make a real contribution in increasing efficacy of drug development pipelines [16] and resulted recently in many new physiological and biochemical insights via biomarkers [2327]. A biomarker is defined as a substance used as an indicator of a biological state. It is characteristic that it is objectively measured and evaluated as an indicator of normal biological processes, nutritional intervention, pathogenic processes, or pharmacological responses to a therapeutic intervention [28] like nutritional intervention (Figure 5).


In this respect molecular mediators in blood plasma [29] are important to study because they can serve as biomarkers to trace people vulnerable and susceptible for metabolic diseases like Obesity and/or IR/DM2. The process to find a proper biomarker is under-estimated and has to follow a long route:
A. The Discovery phase,
B. Qualification phase one, which encompasses:
i. Identification of the biomarker,
ii. Validation analysis,
iii. Proof of the biomarker in a new study,
C. Qualification phase two, which encompasses:
i. Study precision,?
ii. Study robustness,
iii. Study limitation,
iv. Test biomarker in wider population,
D. Finally the application phase in the population is reached [30].
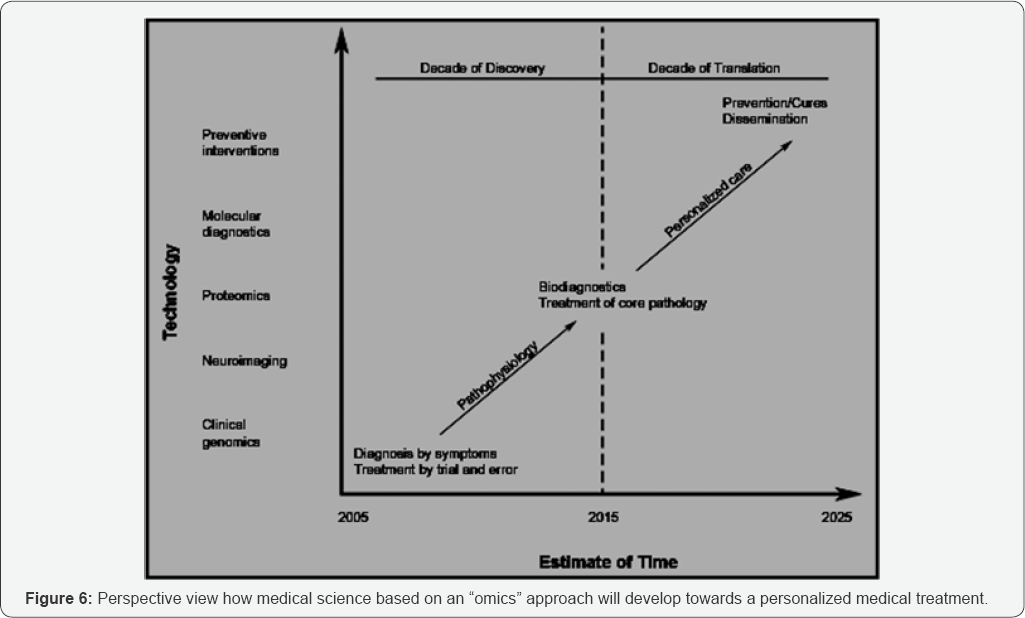
But when an appropriate biomarker for a complex human disease like DM2 is found it can lead to a “Personalized medical treatment” (Figure 6, [19]) were per individual the effects of nutritional intervention should be considered part of a dietary lifestyle strategy to prevent or manage DM2.
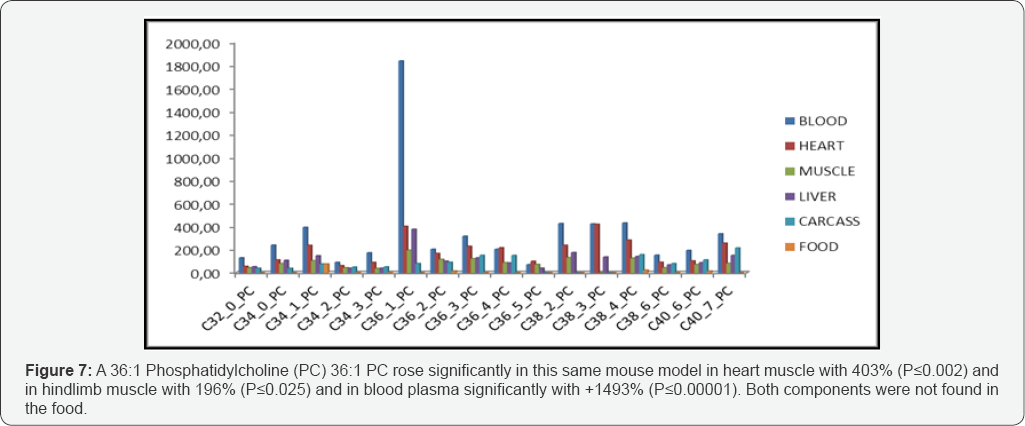
Recently we succeeded to find two potential biomarkers for IR-DM2 in a High Fat diet obese C57Bl/6 mouse model [25]. The first biomarker in this mouse model was a cholesterylester (ChE) -more specific 20:3-ChE- which significantly increased (PP≤00.016) in the fatty heart with 1317% while it rose very significantly (PP≤00.00001) in blood plasma with 1013% in the High-Fat diet group. Meanwhile the 20:3-ChE biomarker (dihomo-y-linolenic; 20:3 n-6) has been classified as a potential DM2 biomarker in a human cohort of the Uppsala Longitudinal Study of Adult Men (ULSAM). The second biomarker was a specific phosphatidylcholine C36:1-PC, which rose significantly (PP≤00.025) mainly in heart muscle with 400% while concentrations increased significantly strongly (PP≤00.002) in blood plasma with 1493% in the High-Fat diet vs. the Cogroup. As an earlier defined prerequisite, both compounds were not found in the food. In all other tissues there were not such clear indications for biomarkers Figure 7.
The 36:1-PC has until this moment not yet been classified in any human cohort study as a potential biomarker "above the disease”. But we found a plausible biochemical modelling related to hypoxic conditions in white adipose tissue which is extensively described in two other manuscripts [25-27]. In addition, the extensive research team of [32] recently described a novel, near-physiological organoid culture system, wherein primary human healthy liver cells form long-term expanding organoids that retain liver tissue function and genetic stability. Recently this in vitro organoid culture system was extended to the propagation of primary liver cancer (PLC) organoids from three of the most common PLC subtypes: hepatocellular carcinoma (HCC), cholangiocarci-noma (CC) and combined HCC/CC (CHC) tumors. In future studies we hope to apply the Systems Biology Lipidomics technique in order to find biomarkers for these forms of liver cancer research for which this recently developed in vitro 3D organoid culture screening technique with human liver cells [32] gives great opportunities.
References
- Rees DA, Alcolado JC (2005) Animal models of diabetes mellitus. Diabet Med 22: 359-370.
- Krook A, O'Rahilly S (1996) Mutant insulin receptors in syndrome of Insulin Resistance. Baillieres Clin Endocrinol Metab 10: 97-122.
- Maassen JA, 'T Hart LM, Van Essen E, Heine RJ, Nijpels G, et al. (2004) Mitochondrial diabetes: molecular mechanisms and clinical presentation. Diabetes 53: S103-S109.
- Nugent DA, Smith DM, Jones HB (2008) A Review of Islet Langerhans Degeneration in Rodent Models of Type 2 Diabetes. Society of Toxicologic Pathology 36(4): 529-551.
- Saltiel AR, Kahn R (2001) Insulin signaling and the regulation of glucose and lipid metabolism. Nature 414: 799-806.
- Salway JG (2004) Metabolism at a Glance. Blackwell Publishing LTD, p.125.
- Salway JG (2006) Medical Biochemistry at a glance, Blackwell Publishing LTD, p.144.
- Marx J (2002) Unraveling the causes of diabetes. Science 296: 686-689.
- Gloyn AL, Mackay DJ, Weedon MN, McCarthy MI, Walker M, et al. (2006) Assessment of the role of common variation in the Transient Neonatal Diabetes Mellitus (TNDM) region in Type 2 Diabetes. A Comparative Genomic and tagging single nucleotide polymorphism approach. Diabetes 55: 2272-2276.
- Gloyn, A.L.; Mackay, D.J.G.; Weedon, M.N.; McCarthy, M.I.; Walker, M.; Hitam, G.; Knight, B.A.;Owen, K.R.; Hattersley, A.T. and Frayling, T.M. (2006). Assessment of the role of common variation in the Transient Neonatal Diabetes Mellitus (TNDM) region in Type 2 Diabetes. A Comparative Genomic and tagging single nucleotide polymorphism approach. Diabetes 55: 2272-2276.
- Sorger PK (2005) A reductionist's systems biology. Current Opinion in Cell Biology 17: 9-11.
- Fontecave M (2010) Understanding Life as Molecules: Reductionism versus Vitalism. Angew Chem Int Ed 49: 4016.
- Friboulet A, Thomas D (2005) Review: Systems Biology-an interdisciplinary approach. Biosensors and Bioelectronics 20: 24042407.
- Ahn AC, Tewari M, Poon CS, Phillips RS (2006) The limits of reductionism in medicine: could systems biology offer an alternative? PLoS Medicine 3(6): e208.
- Kitano H (2002) Systems biology: a brief overview. Science 295(5560): 1662-1664.
- Laaksonen R, Jänis MT, Oresic M (2008) Lipidomics-based safety biomarkers for lipid-lowering treatments. Angiology 59(2 suppl): 65S-68S.
- Keusch GT (2006) What do -omics mean for the science and policy of the nutritional sciences? Am J Clin Nutr 83(suppl): 520S-522S.
- Bain JR, Stevens RD, Wenner BR, Ilkayeva O, Muoio DM, et al. (2009) Metabolomics applied to Diabetes Research moving from information to knowledge. Diabetes 58: 2429-2443.
- van der Greef J, Hankemeier T, McBurney RN (2006) Metabolomics- based biology andpersonalized medicine: moving towards n=1 clinical trials? Pharmacogenomics 7: 1087-1094.
- van der Greef J, Davidov E, Verheij ER, Vogels J, van der Heijden R, et al. (2003) The role of metabolomics in Systems Biology. In Metabolic Profiling: Its Role in Biomarker Discovery and Gene Function Analysis, HARGHigan RGGG (Ed.), p. 170.
- van der Greef J, van der Heijden R, Verheij ER (2004) The Role of Mass Spectrometry in Systems Biology: Data Processing and Identification Strategies in Metabolo-mics. In Advances in Mass Spectrometry, Ashcroft (Ed.), GBAE and Mona-ghan JJ, p. 145.
- Meikle P, Barlow C, Weir J (2009) Lipidomics and Lipid Biomarker Discovery. Australian Biochemist 40: 12-16.
- van Ginneken V, de Vries E, Verheij E, van der Greef J (2016a) Metabolomics in Hind Limb and Heart Muscle of a Mouse model after a High-Fat Diet. Anat Physiol 6: 3.
- van Ginneken VJT, Booms R, Verheij E, de Vries E, van der Greef J (2016b) The Relation between Non-adipose Muscle Fat and Hepatic Steatosis Studied with Localized 1H Magnetic Resonance Spectroscopy (1H MRS) and LCMS Techniques. Anat Physiol 6: 245.
- van Ginneken V, Verheij E, de Vries E, van der Greef (2016c) The discovery of two novel biomarkers in a high-fat diet C56bl6 Obese Model for Non-adipose tissue: a Comprehensive LCMS Study at Hind Limb, Heart, Carcass Muscle, Liver, Brain, Blood Plasma and Food Composition Following a Lipidomics LCMS-Based Approach. Cellular & Molecular Medicine: Open access 2(2): 13.
- van Ginneken V (2017) Targeting Tumor Metabolism: A Biochemical Explanation Related to A Systems Biology Lipidomics Based Approach. J Mol Biomark Diag S: 2.
- van Ginneken V, de Vries E, Verheij E, van der Greef J (2017) [Review]: Potential Biomarkers for "Fatty Liver” (Hepatic Steatosis) and Hepatocellular Carcinoma (HCC) and an explanation of their pathogenesis. Gastroenterol Liver Clin Med 1: 1.
- Stover PJ (2004) Nutritional genomics. Physiol Genomics 16: 161-165.
- Baylin A, Kim MK, Donovan-Palmer A, Siles X, Dougherty L, et al. (2005) Fasting Whole Blood as a Biomarker of Essential Fatty Acid Intake in Epidemiological Studies: Comparison with Adipose Tissue and Plasma. American Journal of Epidemiology 162: 373-381.
- Koulman A, Lane GA, HARGHison SJ, Volmer DA (2009) From differentiating metabolites to biomarkers. Anal Bioanal Chem 394: 663-670.
- Risérus U, Willett WC, Hu FB (2009) Dietary fats and prevention of Type 2 diabetes. Prog Lipid Res 48: 44-51.
- Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, GavARGHó LM (2017) Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med 23(12): 1424-1435.






























