Epigenetic Damage Study, Modification in Methylation and DNA Repair in the Newborn Population of the General Hospital
Martin Noé RC1* and Betzabe MQ2
1 Department of pediatrics and neonatology, General Hospital of Ecatepec, Mexico
2Department of Toxicology of Cinvestav, Mexico
Submission: September 26, 2017; Published: December 20, 2017
*Corresponding author: Martin Noé Rangel Calvillo, Head of pediatrics and neonatology at the General Hospital of Ecatepec, Neonatal pediatric doctor logo. Head of Human milk bank of the same Hospital, Mexico.
How to cite this article:Martin N R, Betzabe M. Epigenetic Damage Study, Modification in Methylation and DNA Repair in the Newborn Population of the General Hospital. Acad J Ped Neonatol. 2017; 6(2): 555735. DOI: 10.19080/AJPN.2017.06.555735
Abstract
Introduction
Methylation is important during embryogenesis, and a correct embryogenic development is susceptible to Epigenetic dysfunction. Some studies have suggested that prenatal exposure to pre-paternal methylation of CPG sites causes changes in DNA repair mechanisms. In the state of Mexico, Ecatepec is considered one of the most polluted sites in terms of pollution. And several previous studies by CINVESTAV have confirmed the presence of contaminants such as benzene, PAHS and PB in the area. In the present work we set out to evaluate the paternal methylation levels of DNA expression gene repair XPC, OGG1 and PARP1 and genetic damage, as well as the ability to repair cell damage in umbilical cord blood obtained in Children of mothers terminated at the General Hospital of Ecatepec. In a period from February to May 2014. We obtained 54 cord samples and also umbilical cord blood samples to assess DNA damage as well as the ability to repair damage at the cellular level by comet study. The results of DNA damage (ORM) were 17.64 +/- 4.95 and showed a marginal positive association with OGG1 (P=0.061) and PARP1 (p=0.087) by exposure at the level of mRNA titers; We observed an inverse relationship of expression in PARP1 paternal titers. The inverse relationship with parents between the ability to repair and the expression of XPC, OGG1 and PARP1 (p <0.05) levels in the OGG1 gene. Where methylation was not correlated with genetic damage with maternal repair ability at levels of gene expression. Our data suggest that the R/N of areas with high levels of environmental pollutants are showing genetic damage. These children with low levels of global methylation are expressed in PARP1 and OGG1 genes that are responsible for repairing proteins that are being affected by the presence of Oxidative contaminants in the environment most changes in Epigenia occur when DNA is methylated, understanding methylation as a function responsible for gene expression and this function responsible for gene expression is regulated by enzymes. During the embryogenesis period methylation is essential to prepare the embryo by inactivating the X chromosome, functions that can be affected by the presence of substances such as benzene, PAHS and Lead (Pb), found in the environment. In this study we evaluated paternal methylation and levels of DNA repair expression in XPC, OGG1, PARP1 genes as well as genetic damage and repair ability in umbilical cord blood cells obtained from term infants born to mothers attended at a hospital General of Ecatepec Dr. JMR.
Objective
Specific: To know if there is epigenetic damage in the population of newborns attended at the general hospital of Ecatepec.
General:
I. To assess the degree of Epigenetic damage in the population of newborns attended at the General Hospital of Ecatepec.
II. Determine the methylation ability in the cells of the selected population under inclusion criteria.
III. Assess by kite study the cellular repair capacity in the selected populations
Material and Method: A total of 44 preterm infants and 60 term newborns from the population attended at the general hospital of Ecatepec were invited to participate in the study. We proceeded on the part of a clinical investigator to take samples of umbilical cord blood which were collected in 2 tubes each containing a volume of 8-9ml, at the same time we proceeded to obtain umbilical cord sample counting with a fragment of 3cm that was included in tube with liquid to transport them. Sixty-one term newborns from the population attended at the general hospital of Ecatepec were invited to participate in the study. Previous consent was obtained from the mother to collect umbilical cord blood in the newborns and the mothers’ blood for Biometric, the breasts also answered a questionnaire to collect information on the history of exposure to pollutants of interest and possible with fusers: consumption of Folic acid and some medications, nutritional status, smoking etc. DNA was extracted from blood cells to determine overall genomic methylation patterns and expression of genes exhibiting differences in the methylation profile.
Method: We selected R/N that met inclusion criteria and previous written authorization from the Mother, we proceeded to apply a questionnaire aimed at detecting possible contact with environmental pollutants. When we counted on the written evidence of this activity, we proceeded on the part of a clinical investigator to take umbilical cord blood samples which were collected in 2 tubes each containing a volume of 8-9ml, at the same time proceeding to obtain sample of Umbilical cord counting with a 3 cm fragment that was included in tube with means of transport, the samples were registered, and delivered to the Researcher in charge of processing them to carry out genetic tests already mentioned. Maternal blood was also extracted to determine hepatic biometry in the mother. And they surrendered to investigator of CINVESTAV for its processing. From the umbilical cord blood sample, monkey nuclear cells were extracted and genetic damage was determined by comet assay as well as cell damage repair capability. Samples were recorded and delivered to the investigator in charge of processing them.
Results
Damage to DNA assessed by comet test: The genetic damage observed in the studied population was very wide (9.11-228.32) with an average of 17.61* - 4.95.
DNA repair capacity: 56% of the samples presented damage repair with an average of 3.99 + - 2-71. Samples than those that did not present repair (44%) were observed a mean of -4.02 + -4.04 Table 1.
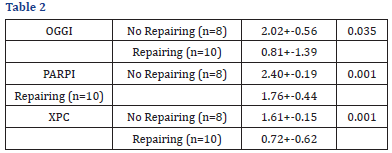
Comparison of genetic damage: We compared samples that presented repair and those that did not present repair with the genetic damage. In those who did not present repair an average of 16.23 +- 2.69 was observed. In those who presented repair a mean of 17.55 +- 5.20 was observed. No significant difference in genetic damage was observed in relation to the samples that presented repair and those that did not present it. Expression levels of OGGI, PARPI and XPC, data obtained from the expression of OGGI and XPC from cord blood showed the highest OGGI gene expression. A marginally significant negative relationship between genetic damage and OGGI expression assessed as the geometric mean was observed and no significant relationship was observed with PARPI or XPC because a lower arithmetic mean value corresponds to a higher level of expression, The umbilical cord samples evaluated that presented higher OGGI expression also presented greater genetic damage. For OGGI a r0.3249 (p=0.0608) was obtained, for PARPI a r=-0.1847 (p=0.3035). For XPC a r=- (P=0.035), p=0.001) and p=0.001 (p=0.001). The results showed that the expression levels (arithmetic mean) of OGGI (p=0.035), PARPI XPC (p=0.001) were significantly higher in the samples that did not present repair compared with those that did present repair, indicating a greater expression of OGGI, PARPI and XPC in the samples that if they repaired the damage.
Conclusion: The genetic damage found in the population studied has a wide range (9.11-28.32) with an average of 17.61. Being the first study performed in this population there are no reference values for comparative analysis, We can say that the R/N of the Municipality of Ecatepec, analyzed have lesions in DNA which makes them susceptible to develop diseases during the course of their life, even adults, especially those who presented greater genetic damage, and this is associated with levels High levels of environmental contaminants in the area. It was also found that 44% of the patient samples analyzed did not present repair which apparently is due to the genetic damage that exceeds the capacity to repair. In the study, a greater genetic damage was observed in umbilical cord samples that presented repair with respect to those that did not present repair of the DNA damage and suggests an altered repair
Keywords: Epigenia; Genetic modifications that are heritages without altering gene sequence; Methylation; Responsible function of gene expression; Epigenetic Dose; Modification in Methylation and DNA repair
Introduction
The statistics in the secretary of health and INEGI have reported an increase in preterm birth, associated with maternal risk factors, have been raised by the current society’s rhythm of life, which among other things exposes us to a large number of contaminants [1-3]. DNA methylation is important for embryogenesis and a suitable embryogenic development is susceptible to an epigenetic dysfunction. During the embryogenesis period methylation is essential to prepare the embryo in differentiation. Specifically, they function by silencing a paternal allele in germinal stem cells, including inactivation of the X chromosome, a crucial activity for the development of the embryo and is given prior to implantation. It is also very important to perform D-methylation of other genes to make the genome available for the developing embryo. The embryo must pass prior to implantation by a global de-methylation and restoration of methylation in the blastocyst stage. Some studies suggest that exposure to contaminations prior to paternal methylation at CPG sites may cause changes in DNA repair mechanisms. The Municipality of Ecatepec is considered the site with the largest population, most contaminated and most industrialized [4-7].
Materials and Methods
44 preterm infants and 60 term newborns from the population served at the general hospital in Ecatepec were invited to participate in the study. We proceeded on the part of a clinical investigator to take samples of umbilical cord blood which were collected in 2 tubes each containing a volume of 8-9ml, at the same time we proceeded to obtain umbilical cord sample counting with a fragment of 3cm that was included in tube with liquid to transport them. Sixty-one term newborns from the population attended at the general hospital of Ecatepec were invited to participate in the study. Previous consent was obtained from the mother to collect umbilical cord blood in the newborns and the mother’s blood for Bh, the breasts also answered a questionnaire to collect information on the history of exposure to pollutants of interest and possible with fusers: consumption of Folic acid and some medications, nutritional status, smoking etc. DNA was extracted from blood cells to determine overall genomic methylation patterns and expression of genes exhibiting differences in the methylation profile.
Method
We selected R/N that met inclusion criteria and previous written authorization from the Mother, we proceeded to apply a questionnaire aimed at detecting possible contact with environmental pollutants.
When we counted on the written evidence of this activity, we proceeded on the part of a clinical investigator to take umbilical cord blood samples which were collected in 2 tubes each containing a volume of 8-9ml, at the same time proceeding to obtain sample of Umbilical cord counting with a fragment of 3 cm that was included in tube with means of transport, the samples were registered, and given to the investigator in charge of processing them to carry out genetic tests already mentioned. Maternal blood was also extracted to determine hepatic biometry in the mother. And they surrendered to investigator of CINVESTAV for its processing Figure 1 & 2.
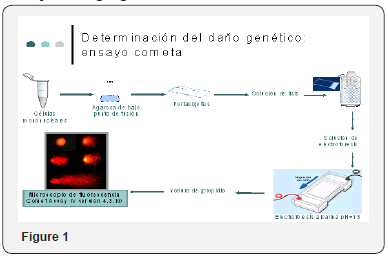
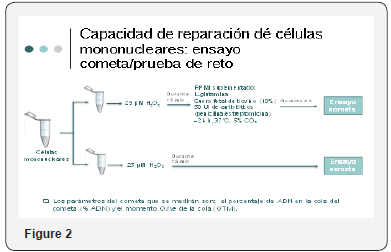
From the umbilical cord blood sample, monkey nuclear cells were extracted and genetic damage was determined by comet assay as well as cell damage repair capability. Samples were recorded and delivered to the investigator in charge of processing them.
Results
Damage to DNA Assessed by Comet Test: The genetic damage observed in the studied population was very wide (9.11- 228.32) with an average of 17.61* - 4.95.
DNA Repair Capacity
56% of the samples presented damage repair with an average of 3.99 + - 2-71. Samples than those that did not present repair (44%) were observed a mean of -4.02 + -4.04.
Comparison of Genetic Damage
We compared samples that presented repair and those that did not present repair with the genetic damage. In those who did not present repair an average of 16.23 + -2.69 was observed. In those who presented repair a mean of 17.55 + -5.20 was observed. No significant difference in genetic damage was observed in relation to the samples that presented repair and those that did not present it. Table 1 Expression levels of OGGI, PARPI and XPC, data obtained from the expression of OGGI and XPC from cord blood showed. Gen measure Geométric interval of trust to 95%. Expression levels of OGGI, PARPI and XPC, data obtained from the expression of OGGI and XPC from cord blood showed the highest OGGI gene expression.

A marginally significant negative relationship between genetic damage and OGGI expression assessed as the geometric mean was observed and no significant relationship was observed with PARPI or XPC because a lower arithmetic mean value corresponds to a higher level of expression, The umbilical cord samples evaluated that presented higher OGGI expression also presented greater genetic damage. For OGGI ar 0.3249 (p=0.0608) was obtained, for PARPI ar=-0.1847 (p=0.3035). For XPC ar=- (P=0.035), p=0.001), and p=0.001 (p=0.001). The results showed that the expression levels (arithmetic mean) of OGGI (p=0.035), PARPI XPC (p=0.001) were significantly higher in the samples that did not present repair compared with those that did present repair, indicating a greater expression of OGGI, PARPI and XPC in the samples that if they repaired the damage.
Discussion
The genetic damage found in the population studied has a wide range (9.11-28.32) with an average of 17.61. Being the first study performed in this population there are no values. Table 2 Indicating a greater expression of OGGI, PARPI and XPC in the samples that if they repaired the damage before dare with H202 12μM. Of reference to perform a comparative analysis, but at the outset we can say that the R/N of the Municipality of Ecatepec, analyzed have lesions in DNA which makes them susceptible to develop diseases during the course of their life, even adults, especially those with the highest Genetic damage, and the above is associated with high levels of environmental contaminants in the area. It was also found that 44% of the patient samples analyzed did not present repair which apparently is due to the genetic damage that exceeds the capacity to repair. In the study, a greater genetic damage was observed in umbilical cord samples that presented repair with respect to those that did not present repair of the DNA damage and suggests an altered repair (Table 3).
Ethical Considerations
This paper does not violate the Helsinki declarations in relation to not causing harm to the participants, and not putting health in the forefront, while maintaining the integrity, and privacy of the participants. It has the authorization of the Ethics and Research Committee of the General Hospital of Ecatepec, who after a rigorous examination authorize the elaboration of this study and it is recorded in proceedings of the Hospital Unit.
Type of Study
Transverse, longitudinal experimental type. With support of technological resources of CINVESTAV and in cooperation with H.G.E. J.M.R. [8-10].
Results
Damage to DNA assessed by comet test
The genetic damage observed in the studied population was very wide (9.11-228.32) with an average of 17.61* - 4.95.
DNA repair capacity
56% of the samples presented damage repair with an average of 3.99 + - 2-71. Samples in which they did not present repair (44%) showed a mean of -4.02 + -4.04 Table 1.
Comparison of genetic damage
We compared samples that presented repair and those that did not present repair with the genetic damage. In those who did not present repair an average of 16.23 + - 2.69 was observed. In those who presented repair a mean of 17.55 + -5.20 was observed. No significant difference in genetic damage was observed in relation to the samples that presented repair and those that did not present it. Expression levels of OGGI, PARPI and XPC, data obtained from the expression of OGGI and XPC from cord blood showed the highest OGGI gene expression. A marginally significant negative relationship was found between genetic damage and OGGI expression assessed as the geometric mean and no significant relationship was observed with PARPI or XPC due To which a lower value of arithmetic mean corresponds to a higher level of expression, the umbilical cord samples evaluated that presented higher OGGI expression also presented greater genetic damage. For OGGI ar 0.3249 (p=0.0608) was obtained, for PARPI one R=-0.1847 (p=0.3035). For XPC a r=-0.0448 (p=0.4085). On the other hand, we compared the samples that presented against those that did not present it, it was observed that the expression levels (Arithmetic Mean) Of OGGI (p=0.035), PARPI (p=0.001) and XPC (p=0.001) were significantly higher in the samples that did not present repair compared with those that did present repair, indicating a higher expression of OGGI, PARPI And XPC on samples that if they repaired the damage [11-20].
Discussion
The genetic damage found in the population studied has a wide range (9.11-28.32) with an average of 17.61. Being the first study performed in this population there are no reference values to perform comparative analysis, but at the beginning We can say that the R/N of the Municipality of Ecatepec, analyzed have lesions in DNA which makes them susceptible to develop diseases during the course of their life even adults, especially those who presented greater genetic damage, and the above is associated with levels High levels of environmental contaminants in the area. It was also found that 44% of the patient samples analyzed did not present repair which apparently is due to the genetic damage that exceeds the capacity to repair. In the study, a greater genetic damage was observed in umbilical cord samples that presented repair with respect to those that did not present repair of the DNA damage and suggests an altered repair. The above results were obtained from samples that were sent to CINVESTAV. 61 cord blood samples and respective cord fragments, the samples were selected by the project managers 43 samples to evaluate expression of the XPC, PARPI and OGG1 genes. They were selected by Master of Science in Toxicology Specialist 44 to evaluate the damage To DNA. And only 18 samples to evaluate the ability to repair the mean maternal age was 24 + - years and the mean maternal hemoglobin concentration was 13.4 + - 1.5g / dl. Most mothers reported having smoked during pregnancy (89.7).Regarding the sex ratio of R/N 28 49%) were male and 30 (51%) were female, 87.2% of the selected were obtained vaginally and the number of pregnancies was variable: 36.2% of the Women were gesta 1. Gestas 2-3 41.4%, and 4 or more deeds 22.4% [21-25].
DNA damage assessed by comet test
The genetic damage observed in the studied population was very wide (9.11-228.32) with an average of 17.61* - 4.95 [26].
DNA repair capability
56% of the samples presented repair of the damage with an average of 3.99 + - 2-71. Samples that did not present repair (44%) observed a mean of -4.02 + -4.04 Table 1.
Comparison of genetic damage
Samples that showed repair and those that did not present repair with the genetic damage were compared. In those who did not present repair an average of 16.23 + - 2.69 was observed. In which Presented repair was observed a mean of 17.55 ± -5.20. There was no significant difference in genetic damage in relation to the samples that presented repair and those that did not present it. Expression levels of OGGI, PARPI and XPC, data obtained from the expression of OGGI and XPC from cord blood showed the highest OGGI gene expression. Expression table of OGGI, PARPI, XPC genes in umbilical cord cells. The mean + standard deviation is presented. N=43. A marginally significant negative relationship between genetic damage and OGGI expression assessed as the geometric mean was observed and no significant relationship was observed with PARPI or XPC because a lower arithmetic mean value corresponds to a higher level of expression, The umbilical cord samples evaluated that presented higher OGGI expression also presented greater genetic damage. For OGGI ar 0.3249 (p=0.0608) was obtained, for PARPI a r=-0.1847 (p=0.3035). For XPC ar=- (P=0.035), p=0.001) and p=0.001 (p=0.001). The results showed that the expression levels (arithmetic mean) of OGGI (p=0.035), PARPI (p=0.001) and XPC (p=0.001) were significantly higher in the samples that did not present repair compared with those who did repair, indicating a higher expression of OGGI, PARPI and XPC in the samples than if they repaired the damage. Relationship between expression levels of OGGI, PARPI and XPC with genetic damage and comparison with resilience. Expression levels of OGGI, PARPI and XPC according to the resuscitation capacity after challenge with H2O2 (12μM) [27-35].
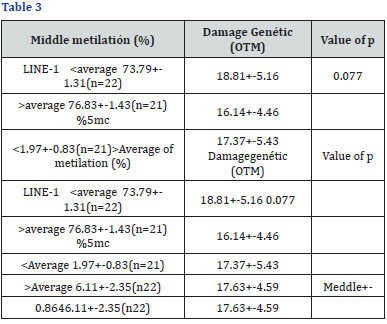
Relationship between global methylation with genetic damage and comparison with repair ability. Further global methylation of DNA measured through the LINE-I marker was significantly associated with a 37% reduction in genetic damage (-0.3725, p=0.01239). In contrast, the overall methylation measured through the% 5- MC was not associated with the presence of genetic damage (r=0.0098 and p=0.9503) nor was there a significant difference in the levels of total methylation (LINE-1 and % 5-mC) in the samples that presented repair compared to those that presented repair. Comparison of global methylation levels with genetic damage [36-38].
Conclusion
1. Genetic damage in the studied population was associated with a lower overall methylation for LINE1
2. High levels of OGG1 expression were positively related marginally with genetic damage.
3. The expression OGG1, PARP1 and XPC of the group that repaired the damage was significantly higher than the group that did not repair.
4. The results suggest that children born in a highly contaminated area such as Ecatepec present genetic damage and alteration in the expression of repair genes, in addition to a decreased overall methylation pattern.
Suggestions
1. Inform the authorities of the health sector of environmental issues.
2. Inform the corresponding authorities about genetic damage found in population.
3. Start local campaign to prevent damage with antioxidant diet
Conflict of interests
We declare that the present study has no conflict of interest whatsoever; Neither the Hospital nor the Author received any gratification during the realization and there is no commitment with any industry that links us with economic interests or different to the scientific medical interest. That the present study was carried out in collaboration with CINVESTAV under previous authorization by the research and teaching ethics committees of the hospital Dr. J.M.R.
References
- Alvarado C, Sánchez GM, Hernández Cadena L, De Vizcaya M, Mujica V et. al (2017) Particulate matter exposure increase gene promoter methylation of DNA repair enzymes in urban children. Mutat Res 81(3): 27-36.
- Gobierno del Estado de México (2010) Instituto Nacional para el Federalismo y el Desarrollo Municipal. En :http://www. estadodemexico.com.mx/portal/ecatepec
- INEGI (Instituto Nacional de Estadística Geográfica e Informática)
(2010) Censo General de Población y Vivienda. En http//www.inegi.
- Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA et. al (2009) Rapid DNA Methylation Changer after exposure to Traffic Particles. Am J Respir Crit Care Med 179(7): 572-578.
- Breton CV, Byun HM, Wenten M, Pan F, Yang A (2009) Prenatal Tobacco Smoke Exposure Affects Global an Gene-specific DNA Methylation. AmJ Respir Crit Care Med 180(5): 462-467.
- Costa V, Quintanilla A, Morada FP (2007) Protein Oxidation, Repair Mechanisms and Proteolysis in Saccharomyces cerevisiae. IUBMB Life 59(4-5): 293-298.
- Reik W, Dean WJ (2001) Epigenetic reprogramming in mammalian development Science. Science 293(5532):1089-1093.
- Gao A, Zuo X, Liu Q, Lu X, Guo W (2010) Methylation of PARP-1 promoter involved in the regulation of benzene-induced decrease of PARP-1 mRNA expression. Toxicol Lett 195(2-3): 114-118.
- Hanova M, Stetina R, Vodickova L, avikova R, Hlavac P (2010) Modulation of DNA repair capacity and mRNA expression levels of XRCCI, hOGGI and XPC genes in styrene-exposed workers. Toxicology and Applied Pharmacology. 248:194-200.
- Lizanti S, Omar WA, Tomaszewski B, De Prins S, Jacobs G (2013) Comparison of Mehods for Quantification of Global DNA Methylation in Human Cells and Tissues PLoS One 8(11): 1-11.
- Valverde M, Rojas E (2009) Environmental and occupational bio monitorin using the comet assay. Mutat Res 681(1):93-109
- Chistmann M, Tomicic MT, RoosWPmkaina B (2003) Mechanisms of human DNA repair: an update. Toxicology 193(1-2): 3-34.
- Alvarado CI, Sanchez G, Pelallo MN, Solís HM , De VA et. al (2013) Oxidative and genetic damage in school children from a heavily polluted city. The toxicologist 389.
- Binkova B, Chvatalova I, Lnenickova Z, Milocova A, Tulupova E et. al (2007) PAH-DNA adducts in environmentally exposed population in relation to metabolic an DNA repair gene polymorphisms. Mutat Res. 620(1-2): 49-61.
- Bollati V, Andrea B (2010) Environmental epigenetics. Heredity (Edinb) 105(1): 105-112
- Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S et. al (2007) Changes in DNA Methylation Patterns in Subjects Exposed to Low-Dose Benzene. Cancer Res 67(3): 876-880.
- Godínez S (2013) Niveles de aductos en el ADN en niños expuestos ambientalmente a benceno y su relación con el dañogenético. Tesis de Maestría. Departamento de Toxicología. Cinvestav.
- Herbstman JB, Tang D , Qu L, Sjodin A, Li Z (2012) Prenatal Exposure to Polycyclic Aromatic Hydrocarbons, Benzo (a)pyrene-DNA Adducts, and Genomic DNA Methylation in Cord Blood Environ Health Perspect. 120(5): 733-738.
- Herbstman JB, Wangs S, Perera F, Lederman SA, Vishnevetsky J (2013) Predictors and consequences of global DNA methylation in cord blood and at three years. PLoS One 8(9): 1-10.
- Jasso PY, Díaz BF, CalderónJ, Yañez L, Carrizales L (2012) DNA Damage and Decreased DNA Repair in Peripheral Blood Mononuclear Cells in Individuals Exposed to Arsenic an Lead in a Mining Site. Biol Trace Elem Res Biological Trace Element Research, 146(2): 141-149.
- Jirtle RL Skinner MK (2007) Environmental epigenomics and disease susceptibility. Nat Rev Genet 8(4): 253-262
- Lic C, Yang X, Xu M, Zhang J, Sun NA (2013) Epigenetic marker (LINE-I promoter ) methylation level was associated with occupational lead exposure. Clin Toxicol (Phila) Clinical Toxicology 51(4): 225-229.
- Likhite N, Warawdekar UM (2011) A Unique Method for Isolation and Solubilization of Proteins after Extraction of RNA from Tumor Tissue Using Trizol. J Biomol Tech Journal of biomolecular Techinques 22(1): 37-44.
- Llop S. BF, Estarlich M, Esplugues A, Rebagliato MC (2010) Preterm birth and exposure to air pollutants during pregnancy. Environ Res 110(8): 778-785.
- Mankamel T, Hokanson R, Fudge R, Chowdharyl R, Busbee D (2006) Altered gene expresión in human cells treated with the insecticide diazinon: corelation with decreased DNA excision repair capacity Hum Exp Toxicol 25(2): 57-65.
- Mendez GJ, Garcia VGG, Lopez CL, Gomez A, Vera E et. al (2008) Genotoxic Effects of Environmental Exposure to Arsenic and Lead on Children in Region Lagunera, Mexico. Ann N Y Acad Sci 1140:358-367.
- Mirbahai L, Kershaw RM, Green RM, Hayden RE, Meldrum RA et. al (2010) Use of a molecular beacon to track the activity of base excision repair protein OGGI in live cells. DNA Repair (Amst) 9(2): 144-152.
- Pavanello S, Bollati V, Pesatori AC, Kapka L, Bolongnesi C et. al (2009) Global and gene-specific promoter methylaton changes are related to anti-BaPDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hidrocarbon-exposed individuals. Int J Cancer 125(7):1692-1697.
- Perera F , Herbstman J (2011) Prenatal environmental exposures, epigenetics, and disease. Reprod Toxicol 31(3):363-373.
- Pilsner JR, Hu H, Ettinger A, Sánchez BN, Wright RO et. al (2009) Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect 117(9):1466-1471.
- Ruchirawat M, Navasumrit P, Settachana D (2010). Exposure to benzene in various susceptible populations: Co-exposures to 1, 3-butadine and PAHs and implications for carcinogenic risk. Chem Biol Interact 184(1- 2):67-76.
- Ruchirawat M, Settachan D, Navasumrit P, Tuntawiroon J, Autrup H (2007) Assessment of potential cáncer risk in children exposed to urban air pollution in Bangkok, Thailand. Toxicol Lett 168(3):200-209
- Sánchez GM, Alvarado CI, Pelayo MN, Monroy PV, Olais R et. al (2012) Environmental exposure to polycyclic aromatic hydrocarbons, benzene, lead, PM10 and PM2.5 and their implications in oxidative damage in Mexican children. The Toxicologist 34.
- Secretaria del, Medio A del, Gobierno del, Distrito F(2008) Inventario de Emisiones de contaminantestóxicos de la ZMVM.
- Singh NP, McCoy MT, Tice RR, Schneider EL (1988) A simple Technique for Quantitation of Low Levels of DNA Damage in Individual Cells. Exp Cell Res 175(1):184-191.
- Szyf M (2011) The implications of DNA methylation for toxicology: toward toxic methylomics, the toxicology of DNA methylation. Toxicol Sci 120(2):235-255.
- Tang WY, Levin L, Talaska G, Cheung YY, Herbstman J (2012) Maternal exposure to polycycli aromatic hidrocarbons and 5- CpG methylation of interferon – e in cord White blood cells. Environmental Health Perspective 120: 1195-1200.
- Wang Q, Ye R, Ye Y, Wan J, Sun P et. al (2012) mRNA Expression Levels Among Cell Regulatory and DNA Damage Genes in Benzene-exposed Worke.
- Wu T, Wang XJ, Tian W, Jaramillo MC, Lau A et. al (2014) Poly (ADPribose) polymerase- I modulates NrF2 dependent transcription. Free Radic Biol Med. 67:69-80.
- Wu YH, Tsai Ch, Cheng YM, Wu TC, Chen CY et. al (2007) Weroderma pigmentosum group C gene expression is predominantly regulated by promoter hypermethylation and contributes to p53 mutation in lung cancers. Oncogene 26(33):4761-4773.
- Yager JW, Eastmond DA, Robertson ML, Paradisin WM, Smith MT (1990) Characterization of micronuclei induce in human Iymphocytes by benzene metabolites. Cáncer Research 50(2):393-399.
- Zawia NH, Lahiri DK, Cardozo PF (2009) Epigenetics oxidative stress, and Alzheimer disease Free Radical Biology an Medicine 46(9):1241- 1249.
- Zhou Z, Lei YWC (2012) Analysis of aberrant methylation in DNA repair genes during malignant transformation of human bronchial epitelial cells induce by cadmium. Toxicol Sci 125(2):412-417.






























