Genetic Landscape of Skeletal Diseases in Pakistan
Saima Siddiqi1*, Azhar Nazir1 and Raheel Qamar2,3
1Institute of Biomedical and Genetic Engineering (IBGE), Pakistan
2Translational Genomics Laboratory, COMSATS University Islamabad, Pakistan
3Pakistan Academy of Sciences, Pakistan
Submission:November 21, 2022; Published: December 12, 2022
*Corresponding author: Saima Siddiqi, Institute of Biomedical and Genetic Engineering Islamabad (IBGE), Pakistan
How to cite this article: Saima S, Azhar N, Raheel Q. Genetic Landscape of Skeletal Diseases in Pakistan. Ortho & Rheum Open Access J. 2022; 20(5): 556049. DOI: 10.19080/OROAJ.2022.20.556049
Mini Review
Pakistan is a country where consanguineous marriages are very common which are manifest due to social and cultural practices in the region. In this country the estimated rate of interfamily marriage is around 60%, due to which recessive mutations in the families (Zafar Iqbal and Hans van Bokhoven, 2014). Recent reports indicate that 16 million people in Pakistan are living with some genetic condition including rare neurological, skeletal and metabolic phenotypes [1]. Skeletal abnormalities characterized by abnormal pattering, defects in linear growth, as well as differentiation and maintenance of the human skeleton with resultant short stature, or dwarfism are together grouped as skeletal dysplasia. This condition constitutes a heterogeneous group of disorders resulting from different genetic variants in humans which together encompass over 450 clinically and genetically heterogeneous conditions involving abnormalities of cartilage and bone [2] and is caused by observable differences in growth patterns, resulting in disproportionate stature between the individuals. It must point out that there is a significant clinical overlap amongst these 450 rare conditions, therefore arriving at a correct diagnosis may require significant medical evaluation and clinical expertise. In addition, National Organization of Rare Diseases (NORD) estimates that there are 300 different genes, mutations in which result in these disorders, which could be inherited in an autosomal recessive, autosomal dominant, X-linked recessive or X-linked dominant [4]. Thus, correct diagnosis requires not only a combination of radiological imaging and clinical assessment but also genetic testing (Duz et al. 2020).
The correct molecular diagnosis of an individual with skeletal dysplasia can lead to improved clinical care, including medical management or treatment based upon personalized care in addition to providing genetic counselling to the families. Correct molecular diagnosis can also lead to eligibility to participate in clinical trials that are underway to targeted therapies for a number of different skeletal dysplasia’s [7].
Next-generation sequencing (NGS) technology has revolutionized both diagnostics and gene discovery in the last few decades, resulting in the identification of disease-causing gene in more than 90% of well characterized skeletal dysplasia. This has resulted in the implementation of genetic testing as a standard component of the diagnostic work-up [2]. Abdullah et al. [8] identified a novel homozygous nonsense variant [c.59 T > A; p.(Leu20*)] in DYM gene to provide the correct diagnosis of Dyggve Melchior Clausen syndrome (DMC, MIM 223800) in patients which opened the door for the identification and correct diagnosis of similar other cases in Pakistani population [8].
A major advantage of NGS technology is that multiple genes can be tested simultaneously [9] using a targeted panel or Whole Exome Sequencing (WES). Many novel genes have also been discovered in the Pakistani population using this technology such as the TMEM251 which is an evolutionary conserved gene, involved in the pathogenesis of chondrocyte differentiation, this disease was first described in Pakistani patients by [10]. While using this technology Umair et al. (2019) reported mutations in 43 genes causing nineteen different Glycogen Storage Diseases (GSDs) in Pakistani population.
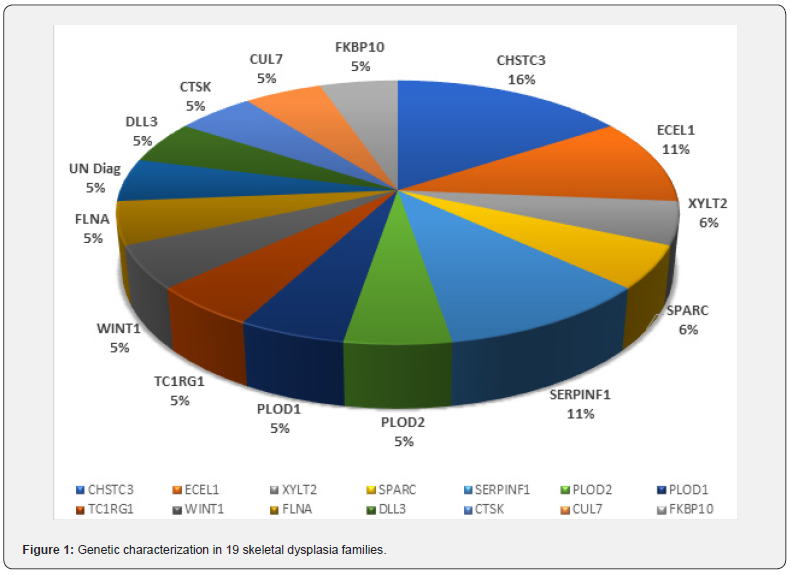
Our Work on 19 skeletal dysplasia families collected from different regions of Pakistan, has also shown the effectiveness of studying index patient from each family by WES analysis. Diagnostic variants in the studied Cohort were identified in 14 genes (Figure 1) in 18 of the families while one family still remains undiagnosed which demonstrates the highly heterogenous nature of skeletal dysplasia in the Pakistani families. In this panel the most common gene was found to be CHST3 that was identified to be the causative factor of the disease in in different 3 families. CHST3-related skeletal dysplasia is characterized by bone and joint abnormalities that worsen over time [10], mutations in this gene reduce or eliminate the activity of the C6ST-1 enzyme. Absence or reduced levels of this enzyme disrupts the normal development of the cartilage and bone, in the affected individual, resulting in the characteristic abnormalities associated with CHST3-related skeletal dysplasia, which is a rare skeletal disorder without known incidence or prevalence. A few years back Duz et al. [3] reported that there were nearly 50 patients with CHST3- related skeletal dysplasia that had been reported worldwide, in whom over 90% of the pathogenic variants were identified in the sulfotransferase domain of the enzyme. Kauser et al. [10] described the genotypic and phenotypic findings in three unrelated families with spondyloepiphyseal dysplasia. This cohort added 15 patients to the CHST3 related dysplasia, since the three families have 8, 5 and 2 affected individuals.
Surgical correction is often only partially successful procedure or available treatment option for CHST3 related patients. Most patients have to undergo multiple procedures by the time they reach adulthood (Unger et al 2010). Surprisingly the work of Seema et al. [11] has shown short statured females also there was the presence of a high frequency of skeletal dysplasia.
Recent breakthroughs in the characterization of mutant proteins, affecting bone tissue homeostasis in rare skeletal diseases, have revealed novel pathways involved in skeletal development and maintenance. The characterization of new dominant, recessive and X-linked forms of the rare brittle bone disease osteogenesis imperfecta (OI) and other OI-related bone fragility disorders was a key player in this advance Besio et al. [12]. The past two decades have contributed a lot in the identification of causative mutations for multiple rare diseases, proving advances in sequencing techniques, to be a very powerful tool in the characterization of the metabolic processes in many organs and tissues [12,13].
Conclusion
Our study suggests that targeted skeletal gene panel sequencing can effectively diagnose up to 90-95% of the skeletal diseases but for the remaining 5-10% of the families we might need Whole exome or Whole genome sequencing. With the use of WES, we have successfully identified 14 causative genes, which are the part of the panel that is commonly used for the genetic diagnosis of skeletal dysplasia. The best strategy for identifying these causative genes may be the use of WES or whole Genome sequencing, which can allow the identification of the genes that have not been reported so far to be involved in skeletal dysplasia.
Pakistani population carry a very high degree of recessive disorders that are comparatively easy to diagnose with NGS data and large families with multiple patients in each family makes the identification easier [14-20].
Availability of data and materials
The datasets generated during and/or analyzed during the current study is available from the author on request.
References
- Dawn News Pakistan (2022) 16m Pakistanis suffering from genetic disorders.
- Scocchia A, Kangas-Kontio T, Irving M, Hero M, Saarinen I, et al. (2022) Diagnostic utility of next-generation sequencing-based panel testing in 543 patients with suspected skeletal dysplasia. Orphanet J Rare Dis 16(1): 412.
- Duz MB, Topak A (2020) Recurrent c.776T>C mutation in CHST3 with four other novel mutations and a literature review. Clin Dysmorphol 29(4): 167-172.
- Longo S, Bollani L, Decembrino L, Di Comite A, Angelini M, et al. (2013) Short-term and long-term sequelae in intrauterine growth retardation (IUGR). J Matern Fetal Neonatal Med 26(3): 222-225.
- Dwan K, Phillipi CA, Steiner RD, Basel D (2016) Bisphosphonate therapy for osteogenesis imperfecta. Cochrane Database Syst Rev 23(7): CD005088.
- Hauer NN, Popp B, Schoeller E, Schuhmann S, Heath KE, et al. (2018) Clinical relevance of systematic phenotyping and exome sequencing in patients with short stature. Genet Med 20(6): 630–638.
- Marzin P, Cormier-Daire V (2020) New perspectives on the treatment of skeletal dysplasia. Therapeutic Advances in Endocrinology and Metabolism 11: 2042018820904016.
- Abdullah, Shah PW, Nawaz S, Shabir Hussain, Asmat Ullah, et al. (2020) A homozygous nonsense variant in DYM underlies Dyggve-Melchior-Clausen syndrome associated with ectodermal features. Mol Biol Rep 47(9): 7083-7088.
- Falardeau F, Camurri MV, Campeau PM (2017) Genomic approaches to diagnose rare bone disorders. Bone 102: 5-14.
- Kausar M, Ain NU, Hayat F, Hunain Fatima, Saad Azim, et al. (2022) Biallelic variants in CHST3 cause Spondyloepiphyseal dysplasia with joint dislocations in three Pakistani kindreds. BMC Musculoskelet Disord 23(1): 818.
- Seema, Abbas S, Ahsan MN, Asghar MS (2022) Frequency of skeletal dysplasia in children with short stature presenting to endocrine clinic: An observational study. J Family Med Prim Care 11(6): 3143-3147.
- Besio R, Chow CW, Tonelli F, Marini JC, Forlino A (2019) Bone biology: insights from osteogenesis imperfecta and related rare fragility syndromes. FEBS J 286(15): 3033-3056.
- Bonthron DT & Foulkes WD (2017) Genetics meets pathology - an increasingly important relationship. J Pathol 241(2): 119–122.
- Ain NU, Muhammad N, Dianatpour M, Marta Baroncelli, Muddassar Iqbal, et al. (2021) Biallelic TMEM251 variants in patients with severe skeletal dysplasia and extreme short stature. Hum Mutat 42(1): 89-101.
- Efthymiou S, Herman I, Rahman F, Najwa Anwar, Reza Maroofian, et al. (2021) Two novel bi-allelic KDELR2 missense variants cause osteogenesis imperfecta with neurodevelopmental features. Am J Med Genet A 185(7): 2241-2249.
- Mortier GR, Cohn DH, Cormier-Daire V, Christine Hall, Deborah Krakow, et al. (2019) Nosology and classification of genetic skeletal disorders: 2019 revision. Am J Med Genet A 179(12): 2393-2419.
- NORD (2021) Dominant Multiple Epiphyseal Dysplasia.
- Paul Saenger, Paul Czernichow, Ieuan Hughes, Edward O (2007) Reiter, Small for Gestational Age: Short Stature and Beyond. Endocr Rev 28(2): 219–251.
- Sabir AH, Morley E, Sheikh J, , Alistair D Calder, Ana Beleza-Meireles, et al. (2021) Diagnostic yield of rare skeletal dysplasia conditions in the radiogenomics era. BMC Med Genomics 14(1): 148.
- Zaka A, Shahzad S, Rao HZ, Kanwal S, Gul A, Basit S (2021) An intrafamilial phenotypic variability in Ellis-Van Creveld syndrome due to a novel 27 bps deletion mutation. Am J Med Genet A 185(10): 2888-2894.






























