In Silico Studies, Comparative Synthesis and Antibacterial Activity of Some Imine Derivatives of Isonicotinic Hydrazide
Francis M Agwom1*, Ezekiel O Afolabi1, Kim J Bot1, Naruka S Yakubu1, Idowu J Olaitan1 and Junior T Kindala2
1Department of Pharmaceutical and Medicinal Chemistry, Faculty of Pharmaceutical Sciences, University of Jos, Nigeria
2Department of Chemistry, Faculty of Sciences, University of Kinshasha, DR Congo
Submission: August 01, 2019; Published: August 29, 2019
*Corresponding author: Francis M Agwom, Department of Pharmaceutical and Medicinal Chemistry, Faculty of Pharmaceutical Sciences, University of Jos, Jos, Nigeria
How to cite this article: Francis M Agwom, Ezekiel O Afolabi, Kim J Bot, Naruka S Yakubu, Idowu J Olaitan, Junior T Kindala. In Silico Studies, Comparative Synthesis and Antibacterial Activity of Some Imine Derivatives of Isonicotinic Hydrazide. Organic & Medicinal Chem IJ. 2019; 8(5): 555747. DOI: 10.19080/OMCIJ.2019.08.555747
Abstract
The yield of synthesis of three derivatives of Isonicotic Hydrazide was compared using refluxing method at 95.C and stirring method at room temperature. The compounds obtained were characterized using IR, H1-NMR and MS spectroscopic techniques and were also assayed for antibacterial activity against Staphylococcus aureus, Escherichia coli, Proteus mirabilis and Klebsiella pneumonia. The possible interactions were studied using PyRx, Arguslab and iGEMDOCK software on bacterial protein targets 3g7e, 1tvf, 3mzd and 4kqr. The results revealed that dimethylamino derivative was obtained with high yield (84%) with stirring at room temperature and the vanillin derivative has the broadest activity across the entire organism tested with an MIC of 150μg against Proteus mirabilis. The docking studies revealed that all the compounds have good possible interaction with target receptors chosen. The Molinspiration and Molsoft bioactivity and drug scores shows that the compounds have good druggability features while the Badapple promiscuity filter identified these compounds as possible promiscuous compounds
Keywords: Docking Imines antibacterial Isonicotinic hyrazide
Introduction
The biological activity of schiff base cut across a range of activities, it is therefore called a versatile Pharmacophore [1]. One of the major activity or the most reported activity of schiff bases is their antibacterial activity [1,2]. Some scholars have also synthesized some analogues of isoniazid imines and tested them against many pathogenic bacteria [3-6]. Another prominent research on the computational studies of imines is that of isoniazid – ketoprofen co-drug imine [7].
With recent growing interest in green chemistry, the stirring method for imine synthesis has been gaining grounds though not as widely as that of microwave. Some Schiff bases of aryl/alkyl aldehydes or ketones with primary amines were synthesized by stirring at room temperature with high to excellent yields in the presence of some catalyst [8]. Worthy of mention is the Schiff base of isonicotinic acid hydrazide (INH) reported by [9].This is because of its closeness to the present study. They carried out the stirring reaction in water and it was shown to perform less than the microwave method though it outperforms the conventional reflux method. This study set out to compare the synthesis of Isonicotinic hydrazide (INH) using stirring and reflux method and to carry out a multi-target and multi-software approach to the computational studies of isonicotinic hydrazide imines.
Materials and Method
Synthesis
Reagents: Isonicotinic hydrazide, Fluka®, Vanillin, Loba Chemie PVT- Ltd®,
4-Dimethylaminobenzaldehyde, Merck®, Piperonaldehyde (heliotropin), BDH Chemicals ltd®
Reflux Method: A 0.005moles (0.6857g) of Isoniazid was completely dissolved in 15ml absolute ethanol and 0.005mole of the Aldehyde (Vanillin, Piperonaldehyde and 4-dimethylaminobenzaldehyde) was dissolved in another 15ml of absolute ethanol and added in drop wise to the Isoniazid solution. Then 4-6 drops of acetic acid were added, and the mixture was refluxed for 3hours at 95⁰C. The reaction was monitored using TLC. The solid crystals obtained was filtered and washed with ethanol.
Stirring Method at Room Temperature: A 0.005moles (0.6857g) of Isoniazid was completely dissolved in 10ml 98% methanol and 0.005mole of the Aldehydes was dissolved in another 10ml of 98% methanol and added in drop wise to the Isoniazid solution with stirring. Then 4-6 drops of acetic acid wereadded, and the stirring was continued for 30 minutes at room temperature. The reaction was monitored using TLC (Scheme 1).
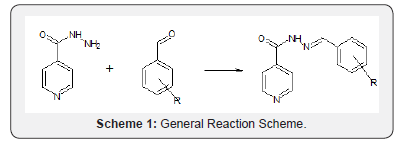
In silico Studies
Docking with PyRx:. PyRx is an open-source software used to perform virtual screening. It uses Vina wizard and Autodock 4.2 as docking softwares. The 3D protein structures of 3G7E (Escherichia coli gyrase B, Co-complexed with prop-2-yn-1-yl{[5- (4-piperidin-1-yl-2-pyridin-3-yl-1,3-thiazole-5-yl)-1H-pyrazol- 3-yl]methyl}Carbamate Inhibitor), 3MZD (Structure of Penicillin Binding Protein 5 from Escherichia coli; cloxacillin acyl enzyme complex), 1TVF (Crystal Structure of penicillin Protein (PBP4) from Staphylococcus aureus) and 4KQR (Crystal Structure of penicillin-Binding Protein 3 from Pseudomonas aeruginosa in complex with (5S)- penicilloic acid) were downloaded from the RCSB Data Bank [10] in pdb format and saved for loading into PyRx software [11].
The most probable binding pocket of each protein structure was identified using the Computed Atlas of Surface Topography of Proteins (CASTp) platform [12]. The amino acids representing the binding site of the Protein target were selected and the grid box adjusted to set the Vina Search space. The grid box was set big enough to allow the ligand to move freely in the selected search area. The Vina calculation was then allowed to run.
Docking with iGEMDOCK 2.1: The 3D protein structures of 3G7E, 3MZD, 1TVF and 4KQR were downloaded from the Worldwide Protein Data Bank in pdb format and saved for loading into iGEMDOCK software. The prepared protein was loaded into iGEMDOCK platform and the binding site type set as by bounded ligand. The center of binding site was defined as MOL, the size of the binding site was set at 10 Å and the hydrophobic preference and Electrostatic preference were both set at 1.0 docking scoring functions. The Docking Accuracy Settings (GA parameters) were set as follows: Population size- 200, Generations- 70, Number of Solutions – 2. Standard Docking was selected, and the settings were submitted for docking calculations and the Results were saved and analyzed.
Docking with Arguslab 4.0: The binding sites for 3G7E and 3MZD were obtained from CASTp and the amino acid residues were selected and made into a binding site group in Arguslab. For 4KQR and 1TVF the amino acid residues of the already extracted cavity were all selected and made into a binding site group in Arguslab. The ligands were also made into a ligand group and flexible Argusdock docking calculations were carried out. The results were analyzed and viewed using Discovery Studio software.
Calculation of Molecular Properties and Bioactivity
Scores: The Molsoft and Molinspiration platforms were used.
Promiscuity Filtering of Compounds: This was carried out using the BADAPPLE platform [13].
Antibacterial Assay (Disc Diffusion Method)
The bacteria (Staphylococcus aureus, Escherichia coli, Proteus mirabilis and Klebsiella pneumonia) were seeded into four agar plates for each organism, for the compounds DI, PI, VI and INH. Each of the seeded plate was divided into six sections representing the five concentrations of each compound and the solvent control (DMSO). Autoclaved filter paper discs were introduced into each concentration of each sample as well as the solvent control.
After soaking for sometimes one filter paper disc is picked and carefully placed in the corresponding position on the already labelled agar plates. The plates were then incubated for 24h at 37⁰C in the oven. After incubation the result was read by measuring the zone of inhibition in millimeter.
Result and Discussion
Spectroscopic Data’
Compound 1: H1-NMR (400MHZ Bruker Instrument, DMSO-d6) –δ ppm, 8.79(2H), 8.356(2H), 7.84(1H), 7.36(1H), 7.11(1H), 6.86(1H) and 3.84(3H); IR (Shimadzu, KBr) - Wavenumber cm-1 3055-400(bs), 1790(w), 1651(m), 1566(m), 1512(m), 3425(w), 1427(w) [bs – broad and Strong, w – weak, m – medium]; High-resolution Mass Spectra (amazon SL Bruker Instrument, ESI) - m/z (molecular ion peaks) 272(+1nH), 294 (+1nNa), 565.1(+8nNa) Molecular formula obtained: C14H13N3O3
Compound 2: IR(Shimadzu, KBr) – Wavenumber cm-1 3649(s),2885(m), 2816(m), 2252(w), 1859(vw), 1790(vw), 1666(m), 1573(m),1435(m) [s – strong, m- medium, w – weak, vw – very weak]; High-resolution Mass Spectra (amazon SL Bruker Intrument, ESI) – m/z (Molecular ion Peaks) 270.03(+1nH), 292.01(+1nNa), and 561.07(+8nNa) Molecular Formula Obtained: C14H11N3O3
Compound 3: H1-NMR (400MHZ Bruker Instrument, DMSO-d6) –δ ppm, 8.76(2H), 8.32(2H), 7.78(1H), 7.59(1H), 7.55(1H), 6.80(1H), 6.75(1H), 3.32(6H); IR (Shimadzu, KBr) – Wavenumber cm-1 3402(w), 2885(m), 2808(m) 1790(w), 1697(w), 1527(w), 1411(w) [w- weak, m- medium]
High-resolution Mass Spectra (amazon SL Bruker Instrument, ESI) – m/z (molecular ion peaks) 269(+1nH), 291.1(+1nNa), 559.2(+8nNa) Molecular formula obtained: C15H16N4O
All the synthesis worked well with the aldehydes using the reflux method except for Piperonaldehyde (Compound 2) where the yield was low. On the other method of stirring used, the reaction did not work with the vanillin (Compound 1) while the Dimethylamino benzaldehyde (Compound 3) works well with both methods (Table 1). with a slightly higher yield observed with the stirring method. However, this observation differs a little with some other work where the stirring was shown to work better than the reflux method using aldehydes containing hydroxyl groups [14] just like that in the vanillin, though some of the aldehydes used in that study also contain halogen electron withdrawing groups alongside the hydroxyl group [14].
It was also observed that the imine products obtained are all yellow in color though they only differ in terms of the shades of yellow color. The starting materials utilized for the synthesis are all white powders and crystals therefore the change in color shows that a likely chromophore has been formed given the product it characteristic color. This corresponds with several other researches where Schiff bases are reported as yellow in color [6,15,16]. This could also explain the reason dimethylamino benzaldehyde is used as reagents for derivatization and identification of alkaloids and saponins in plant samples [17] (Figures 1 & 2).
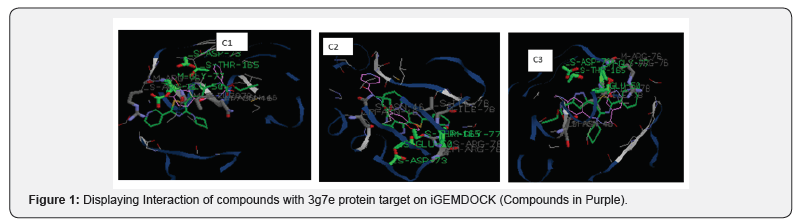
For the iGEMDOCK, the binding pocket used is that of the co-crystalised ligand of the protein. The iGEMDOCK binding affinities calculation of the compounds on 3g7e protein (Table 2) reveals that the binding energy of Compound C3 is more than that of the others. It also shows the major interaction with receptor is via Vander Waal’s interaction than Hydrogen bonding coming from the benzene ring and pyridine ring. Electrostatic interactions are absent as expected because of the absence of charges in theirchemical structures. The amino acids residues of the protein involve in the interaction with each of the compounds is displayed in Figures 1 & 2. Careful examination of the interaction shows that compound C2 interact mainly with glycine, arginine, aspartate and Glutamate while C1 and C3 shows additional interaction with Threonine165. The overall number of the amino acid residues around C3 is less than that of the others and that explains why it has a higher energy, thus implying less activity (Table 3).
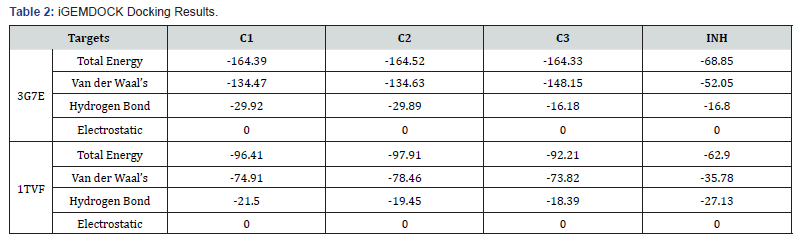
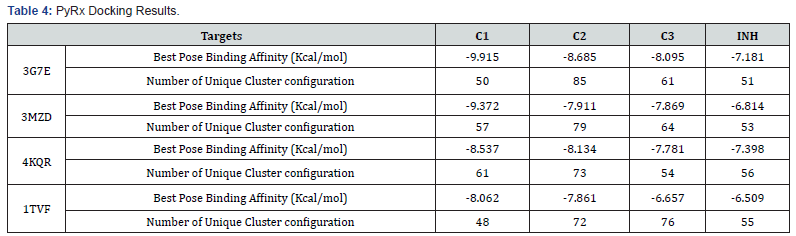
The results of the compounds on the same target protein( 3g7e) in PyRx did not corroborate that of the iGEMDOCK analysis showing that compound C1 and C2 have higher energy and hence less favorable compared to C3, although the difference is not very significant. The similarity in binding energy and pose for C1 and C2 could be due to their close resemblance in chemical structures. The C2 is just one bond away from C1 (i.e. bonding the methoxy and hydroxyl group of C1 gives C2), thus C2 will have some level of rigidity compare to C1.
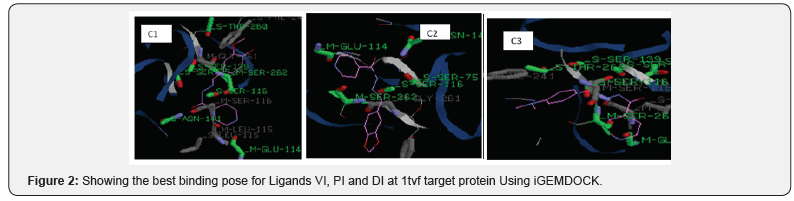
A different trend can be observed in 1tvf target on iGEMDOCK. The interaction analysis reveals that interaction of the compounds with the receptor (1tvf) is mainly via the pyridine ring (see figure 2) and the main amino acid residue involved with the interaction is serine. Looking at the structure of serine it has four hydrogen bond donors [18] while the main hydrogen bond acceptors on the compounds come from the pyridine portion (Figures 3-6).
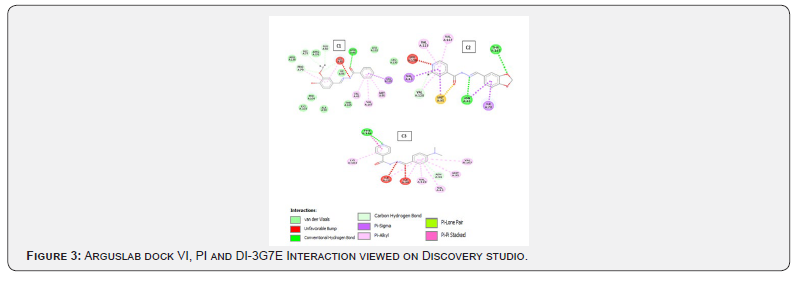
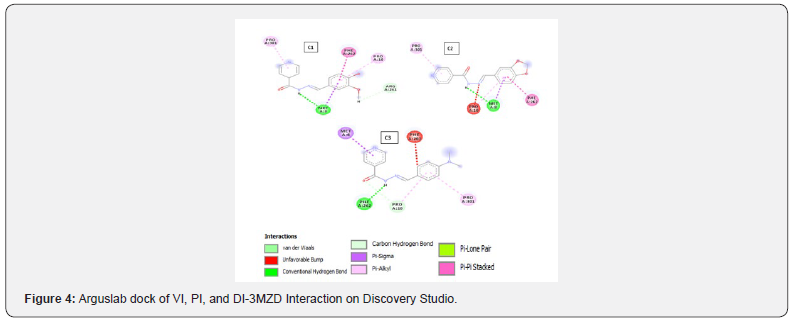
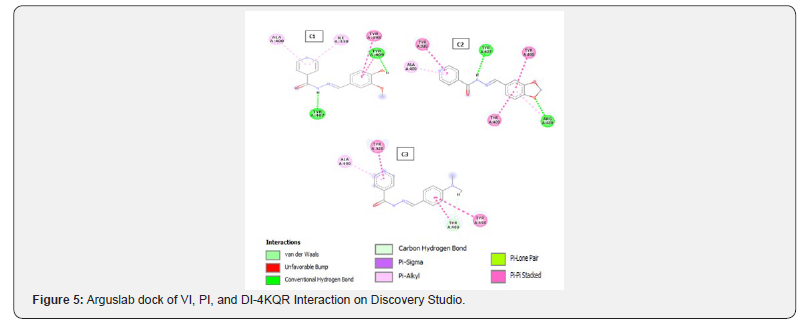
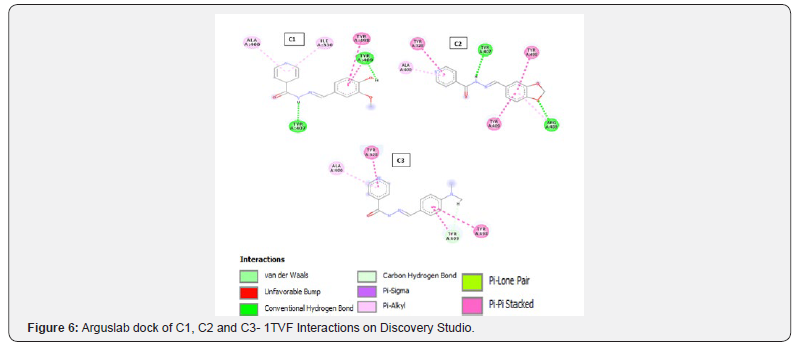
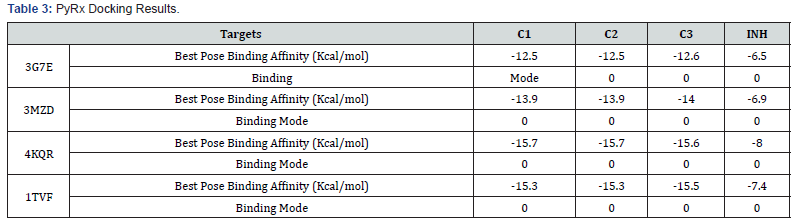
The results of all the compounds docked in the four targets using Arguslab showed a similar trend to iGEMDOCK and on target 4kqr on PyRx. The compound C1 has the best binding affinity followed by compound C2. Compound C3 has the least binding affinity compared to C1 and C2 in most of the targets used (Table 4). This trend also conforms to that observed in the biological assay as C3 showed least activity experimentally. All the compounds (C1, C2 and C3) have better affinities compared to the unsubstituted INH which implies that the modification on INH might likely improve it antibacterial activity. However, looking at the interactions of the compounds with the binding site, it can be said that theoretically the imine functionality only slightly take part in the interaction with the chosen targets (Tables 5-7).
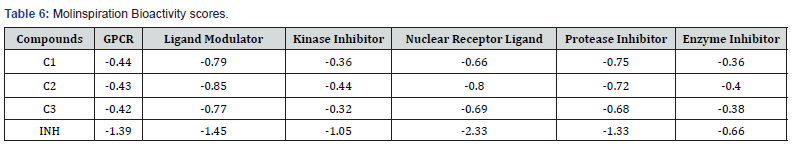
In all the docking studies the compounds showed some promising interactions with negative binding energies. Therefore, if the compounds could have direct access to these protein targets some form of inactivation of the bacteria could be observed. Drug score values indicate overall potential of a compound to be a drug candidate. Mol inspiration is a web-based tool used to predict the bioactivity score of the synthesized compounds against regular human receptors such as GPCRs, ion channels, kinases, nuclear receptors, proteases and enzymes as presented in Table 6.
The interpretation of the results is such that molecule having bioactivity score more than 0.00 is most likely to exhibit considerable biological activities, while values 0.50 to 0.00 are expected to be moderately active and scores less than 0.50 is presumed to be inactive. The results as presented in Table 6 clearly shows that synthesized imines derivatives are less likely to be active at those receptor sites of kinase, Enzyme inhibitor, and Nuclear receptors but are moderately likely to be active at GPCR, Protease, Enzyme Inhibitor. The bioactivity score of compounds is suggestive of moderate interaction with all drug targets. These imines are being positioned for antibiotic activity a strong activity on those receptor or multiple receptor sites are not desirable features since those receptors are the human receptors, therefore in pursuanceof selectivity for bacteria targets and safety all the compounds can be categorize as satisfactory. However, in comparison to the INH it turns out to have better selectivity as it is inactive in all the receptors. These results differ significantly with those of imidazoles and benzimidazole- carbamates reported by other researchers [19-22].
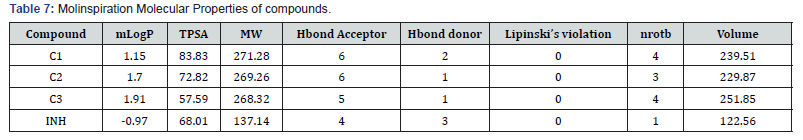
The molecular properties of the compounds as calculated by molinspiration showed all the compounds obey the Lipinski’s rule of five which is an indication of druggability. The number of hydrogen bond acceptors/donors, number of rotatable bonds and surface area and volume are properties of the compound that affect it binding to receptors. For example, all the compounds have higher Hydrogen bond acceptors than donors; therefore, they are more likely to interact with amino acid residues at receptor sites that possess good hydrogen bond donor groups for hydrogen bonding.
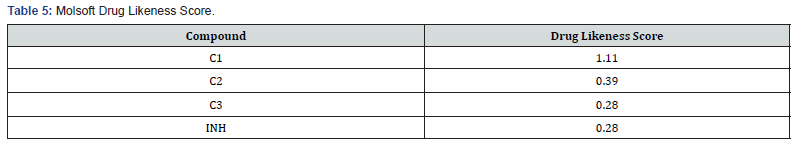
Since it has been reported that the origins of recently launched drugs were mostly derived by modification of known drug structuresor from lead structures obtained from the scientific literature [23], it is therefore important to explore the druggabbility of design molecules relative to those in use. The molsoft software gives that kind of predictions. As presented in Table 5 the compound C1 has the highest druggability value making it a possible lead like compound.
The Badapple software provides a simple method for the rapid identification of likely promiscuous compounds using associated scaffolds. Badapple generates a score associated with a pragmatic, empirical definition of promiscuity, with the overall goal to identify “false trails” and streamline workflows [13]. The result presented in Table 8 shows that the Badapple software has identified the pyridine as the worst offending scaffold giving the compounds a high promiscuity score. Though the importance of the pyridine ring in medicinal chemistry is widely established [24] caution should also be exercised when designing compounds containing the pyridine as it has also been identified as a major scaffold for promiscuity [25].
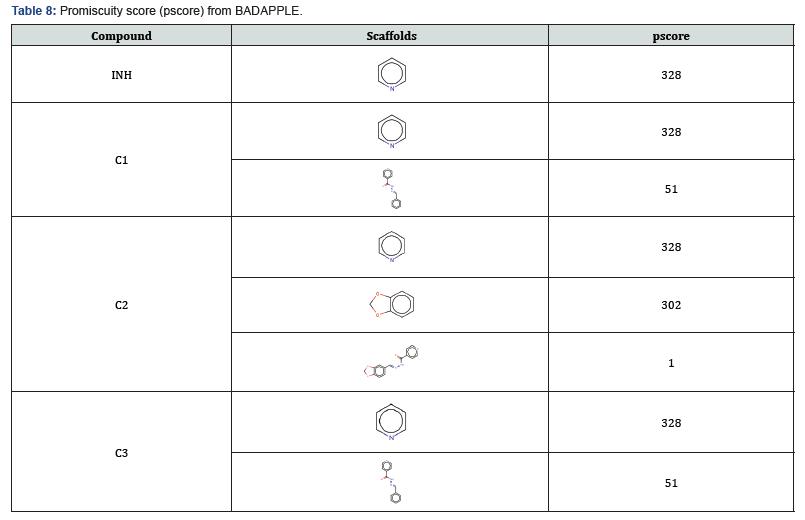
Overall, the promiscuity score of the compounds is considered high. Base on the antibacterial activity in Table 9 Compound C2 showed activity in all the organisms tested (Proteus mirabilis, E. coli, Staph aureus and Klebsiella pneumonia) though the activity is very minimal with the best minimum Inhibitory concentration (MIC) of 150μg for both E. coli and P. mirabilis. The activity of compound C1 is also across the four organisms, (i.e. similar spectrum of activity) but its activity is obviously less than that obtained with C2 in terms of potency. Its best MIC is seen for Proteus at 150μg/ ml. On the other hand, Compound C3 is the poorest of them all showing no activity in all except for E. coli signifying that it has narrow spectrum of activity. Comparing the compounds with the INH it shows that the INH has broad spectrum of activity and better potency than all the synthesized compounds with much wider zone of inhibition and as well as the best MIC for Staphylococcus aureus at 50μg/ml.

Several similar imine compounds have been shown to possess similar spectrum of activity as the one obtained for C2 and C1 [18]. For example, in 2008 Jesmin et al. [26] reported a similar MIC to Staphylococcus of an imine compound they synthesized. The activity of the isoniazid reported here disagrees a bit with that of Ladishav et al., though different strains of organisms were used. There has been some activity of isoniazid previously reported against E. coli and Staphylococcus aureus [27,28].
Conclusion
The best method of synthesis depends on the nature of the aldehyde. Though all the three compounds showed favorable computational properties for lead-likeness, however, experimental activity were not impressive.
Acknowledgement
The authors acknowledge the African Center of Excellence in Phytomedicine Research and Development (ACEPRD), University of Jos, for financial support.
References
- Kajal A, Bala S, Kamboj S, Sharma N, Saini V (2013) Schiff Bases : A Versatile Pharmacophore. Journal of Catalyst 1–14.
- Pawaiya A, Pawaiya P, Rajput S, Agrawal M C, Bhatnagar R K (2014) Synthesis, characterization and antifungal activity of hydrazone schiff base. International Journal of Current Research 6(9): 8423–8426.
- Chohan Z H, Arif M, Shafiq Z, Yaqub M, Claudiu T (2008) In vitro antibacterial, antifungal & cytotoxic activity of some isonicotinoylhydrazide Schiff â€TM s bases and their cobalt ( II ), copper ( II ), nickel ( II ) and zinc ( II ) complexes. Journal of Enzyme Inhibition and Medicinal Chemistry 21(1): 95–103.
- Habala L, Varényi S, Bilková A, Herich P, Valentová J, et al. (2016) Antimicrobial Activity and Urease Inhibition of Schiff Bases Derived from Isoniazid and Fluorinated Benzaldehydes and of Their Copper(II) Complexes. Molecules 21(1742): 1–14.
- Singh R, Kashaw S K, Mishra V K, Mishra M, Rajoriya V (2018) Design and Synthesis of New Bioactive 1,2,4-Triazoles, Potential Antitubercular and Antimicrobial Agents. Indian Journal of Science 80(1): 36–45.
- Yong J N, Mainsah E N, Ntum S E, Ndifon P T (2016) Synthesis, Characterization and Antibacterial Studies of Some Isoniazid-derived Schiff Bases. International Research Journal of Pure and Applied Chemistry 12(1): 1–8.
- Rehman N, Khalid M, Bhatti M H, Yunus U, Albert A, et al. (2018) Schiff base of isoniazid and ketoprofen : synthesis, X-ray crystallographic, spectroscopic, antioxidant, and computational studies. Turkish Journal of Chemistry 42:639–651.
- Ali E, Naimi Jamal M R, Dekamin M G (2013) Highly efficient and rapid synthesis of imines in the presence of nano-ordered MCM-41-SO 3 H heterogeneous catalyst. Scientia Iranica 20(3): 592–597.
- Thomas A B, Sharma P A, Deshpande A D, Tupe P N, Badhe R V, et al. (2009) Green route synthesis of Schiff bases of isonicotinic acid hydrazide. Green Chemistry Letters and Reviews 2(1): 23–27.
- Research Collaboratory for Structural Bioinformatics (n d) RCSB Protein Data Bank
- Dallakyan S, Olson A J (2014) Small-Molecule Library Screening by Docking with PyRx. Methods Mol Biol 1263: 243–250.
- Binkowski T A, Naghibzadeh S, Liang J (2003) CASTp: Computed Atlas of Surface Topography of proteins. Nucleic Acids Res 31(13): 3352–3355.
- Yang J J, Ursu O, Lipinski C A, Sklar L A, Oprea T I, et al. (2016) Badapple : promiscuity patterns from noisy evidence. Journal of Cheminformatics 8(29): 1–14.
- Rao V K, Reddy S S, Krishna B S, Mohan K R, Raju C N, et al. (2010) Green Chemistry Letters and Reviews Synthesis of Schiff â€TM s bases in aqueous medium : a green alternative approach with effective mass yield and high reaction rates. Green Chemistry Letters and Reviews 3(3): 217–223.
- Bhagat S, Sharma N, Chundawat T S (2013) Synthesis of Some Salicylaldehyde-Based Schiff Bases in Aqueous Media. Journal of Chemistry 1–4.
- Nair M S, Arish, D, & Johnson, J (2016) Synthesis, characterization and biological studies on some metal complexes with Schiff base ligand containing pyrazolone moiety. Journal of Saudi Chemical Society 20: S591–S598.
- Le A V, Parks S E, Nguyen M H (2018) Improving the Vanillin-Sulphuric Acid Method for Quantifying Total Saponins. Technologie 6(84).
- Abdel hamid M K, Mccluskey A (2014) In Silico Docking, Molecular Dynamics and Binding Energy Insights into the Bolinaquinone-Clathrin. Terminal Domain Binding Site Molecules 19: 6609–6622.
- Husain A, Ahmad A, Alam S, Asif M, Bhutani R, et al. (2016) Synthesis, molecular properties, toxicity and biological evaluation of some new substituted imidazolidine derivatives in search of potent anti-inflammatory agents. Saudi Pharmaceutical Journal 24: 104–114.
- Khan T, Dixit S, Ahmad R, Raza S, Azad I, et al. (2017) Molecular docking, PASS analysis, bioactivity score prediction, synthesis, characterization and biological activity evaluation of a functionalized 2-butanone thiosemicarbazone ligand and its complexes. Journal of Chemical Biology 10(3): 91-104.
- Raghunath M, Singh A, Viswanathan C L (2015) Molecular Descriptors and Bioactivity Scores of 6- Substituted Benzimidazole-2-Carbamates as Potential Anticancer Agents. World Journal of Pharmacy and Pharmaceutical Sciences 4(4): 1438–1445.
- Singh S, Gupta A K, Verma A (2013) Research Journal of Pharmaceutical, Biological and Chemical Sciences Molecular Properties and Bioactivity score of the Aloe vera antioxidant compounds – in order to lead finding. Research Journal of Pharmaceutical, Biological and Chemical Sciences 4(2): 876–881.
- Proudfoot J R (2002) Drugs, leads, and drug-likeness : an analysis of some recently launched drugs. Bioorganic and Medicinal Chemistry 12(12): 1647–1650.
- Hamada Y (2018) Role of Pyridines in Medicinal Chemistry and Design of Inhibitors Possessing Possessing a Pyridine Pyridine Scaffold In IntechOpen.
- Bruns R F, Watson I A (2012) Rules for Identifying Potentially Reactive or Promiscuous Compounds. Journal of Medicinal Chemistry 55(22): 9763-9772.
- Jesmin M, Ali M, Salahuddin M S, Habib M R, Khanam J A (2008) Antimicrobial Activity of Some Schiff Bases Derived from Benzoin, Salicylaldehyde, Aminophenol and 2,4 Dinitrophenyl Hydrazine. Mycobiology 36(1): 70–73.
- Rosner J L (1993) Susceptibilities of oxyR Regulon Mutants of Escherichia coli and Salmonella typhimurium to Isoniazid. Antimicrobial Agents and Chemotherapy 37(10): 2251–2253.
- Tritz, G J (1974) Protection of Escherichia coli from Isoniazid Inhibition Antimicrobial Agents and Chemotherapy, 5(3), 217–222.






























