Qsar Study of Derivatives for Antifungal Evaluation of Novel Benzimidazole Type of Fluconazole Analogues Invoking Quantum Mechanical Descriptors
Seema Dhail1* and TanmoyChakraborty2
1Department of Science, Manipal University, India
2Department of Chemistry, Manipal University, India
Submission: May 18, 2018; Published: May 30, 2018
*Corresponding author: Seema Dhail, Department of Science, Manipal University, India, Email: seemaresearch21@gmail.com
How to cite this article: Seema D, Tanmoy C. Qsar Study of Derivatives for Antifungal Evaluation of Novel Benzimidazole Type of Fluconazole Analogues Invoking Quantum Mechanical Descriptors. Organic & Medicinal Chem IJ. 2018; 7(1): 555702. DOI: 10.19080/OMCIJ.2018.07.555702
Abstract
Recently Hui Zhen Zhang et al. reported a series of benzimidazole type of fluconazole. They synthesized and characterized them by 1H NMR, 13C NMR, IR, MS and HRMS spectra. An antimicrobial activity of all the newly synthesized compounds has done in in vitro conditions by using two-fold serial dilution technique. Compound named as 3,5-bis(trifluomethyl)phenyl benzimidazoles gave comparable and even stronger antifungal efficiency in comparison with reference drugs such as Chloromycin, Norfloxacin and Fluconazole. They synthesized 27 analogs of benzimidazole. In this report, we have tried a qualitative correlation of antifungal activity of a series of benzimidazolein terms of their computed DFT based global quantum mechanical descriptors. The effects of substitutions on the experimental activities of the instant compounds are explained invoking Global descriptors. Local descriptors have been used to mention site selectivity and mechanistic pathways of bio-active molecules. Finally, QSAR models were derived by means of k nearest neighbours and by Genetic Programming (GP) approaches
Abbrevations:GP: Genetic Programming; QSPR: Quantitative Structure Property Relationship
Introduction
Benzimidazole is a combination of benzene and imidazole ring containing nitrogen, oxygen sulphur and its derivatives. Benzimidazole regarded as one of the promising class of bioactive heterocyclic compounds. It exhibits wide range of biological activities such as anti-microbial, anti-viral, anti-diabetic, anti-cancerous activity, numerous anti-oxidants, anti-parasitic, anti-helminthic, anti-proliferative, anti-HIV, anti-inflammatory, anti-hypertensive, anti-neoplastic, proton pump inhibitor etc [1-4]. Benzimidazole is having electron richer properties as compare to imidazole or triazole due to its larger conjugated structure having fused ring of imidazole with benzene. Fluconazole is a triazole antifungal drug used for treating superficial and systematic fungal infections against Candida albican sand Cryptococcus neoformans It is considered that Fluconazole restrain the biosynthesis of ergosterol and thus inhibiting the growth of fungi due to the triazole ring which efficiently coordinate with iron(II) ion of heme. In spite of this action generally it leads to severe toxicity and precludes its application in the treatment of deep-seated mycoses and life threatening systemic infections [5]. Due to its increasing resistance, narrow antifungal spectrum and even low activity against non-Candida fungi more efforts are needed towards further researchers of Fluconazole for developing more effective antifungal agents with broader antimicrobial spectrum and better therapeutic indexes.
With the increased knowledge of science, new field is introduced named as computational chemistry which is useful for changing or modifying the structures of drugs or chemicals to make new and effective drugs to overcome such a big issue. According to central idea of chemistry any physical, chemical or biological activity of a molecule is depend on its geometrical and topological structure because all the information is coded in its structure only [6,7]. All the chemical interaction and reactions between any two molecule is specific because all the sites of a molecule are not active i.e. the reactions takes place at specific site not globally. The challenging task of chemist is to correlate/evaluate the structure with its property/activity of selected molecule. Quantitative Structure- Activity Relationship (QSAR) and Quantitative structure- Property Relationship (QSPR) is the field of science which is engaged in correlating and evaluating the property/activity of any selected compound depending on its structure. This scientific approach helps in conserving the resources and also accelerates the process of new molecules development for use as drugs, materials, additives, or for any other purpose. According to laws of quantum mechanics molecules are objects..
Generally theoretical descriptors and experimental descriptors were used and theoretical are defined and derived on the basis of quantum mechanics and, at the same time, there are experimental descriptors too [8,9]. But according to research findings it was proven that quantum mechanical descriptors are much superior to the experimental descriptors because in quantum mechanical descriptors there is no statistical dispersion. The site selectivity of selected molecule can only be defined by various DFT descriptors like local softness, local hardness and fukui functions [8].
Initially QSAR methods were developed by Hansch and Fujita [10]. The QSAR paradigm has been successfully applied in vast number of areas like in designing many drugs as well as agrochemical compounds. Besides its simplicity and speed, QSAR has advantages to predict some transport and metabolic processes which occur once the compound is administered [11]. In present research work, we have tried to explore a correlation between the known antifungal activities of benzimidazole derivatives in in vitro conditions with their global as well as with the local quantum mechanical descriptors. Further using local density descriptors we try to locate the most reactive site in the drug molecules.
Finally, k Nearest Neighbors and Genetic Programming was used for QSAR modeling.
Computational Methods
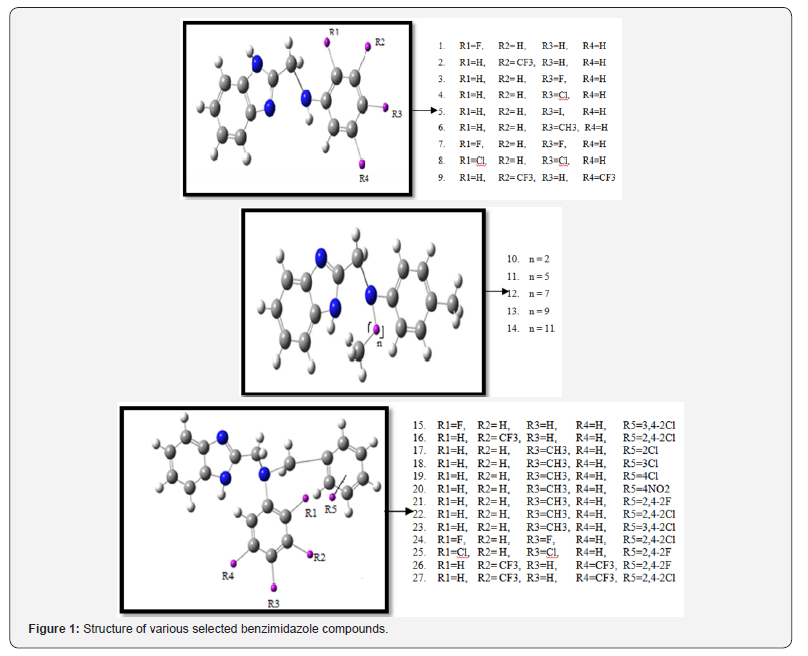
In this venture, we have studied twenty seven derivatives of novel benzimidazole type of Fluconazole analogues which are used in the treatment of fungal infections caused by Candida albicans, Saccharomyces cerevisiae, and A. flavus [12]. The parent structure of novel benzimidazole and its derivatives which have been used in this study has been presented in the Figure 1. The structures of all novel benzimidazole derivatives have been created by ISIS Draw 2.2 software and 3D modeling of the instant compounds has been performed with the help of Gaussian 09 software. We have evaluated global descriptors using semi empirical using PM3 method. Gaussian 09 software have been used to calculate the global descriptors by using the DFT based descriptors. It may be pointed out that the semi empirical methods are more reliable than ab-initio methods [13] in QSAR/QSPR study. Invoking Koopmans’ theorem [7,14] ionization energy (I) and electron affinity (A) have been calculated using formulae


Thereafter, using I and A, the electronegativity (χ), global hardness (η), molecular softness (S) and electrophilicity index (ω) have been computed. For the computation of global reactivity descriptors, we have used the following equations-

Where, μ represents the chemical potential of the system



For the computation of local descriptors, we have used the PM3 method [7,18-20]. The HOMO and LUMO eigen functions obtained from PM3 calculation have been used for computing fukui function (f- , f+,f0) with the help of following formulae [3,6]:
For governing electrophilic attack,

For governing nucleophilic attack

For governing radical attack

The local softness values have been evaluated using the following equations: [16,24-27]
For governing electrophilic attack

For governing nucleophilic attack

For governing radical attack

The local philicity indices have been evaluated through the equations as follows: [6]
The ω- is for governing electrophilic attack

The ω+ is for governing nucleophilic attack

The ω0 is for governing radical attack

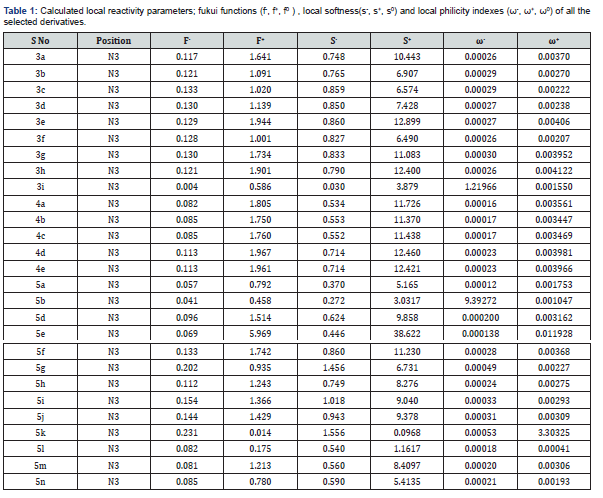
The observed activity and as well as reactivity parameters, calculated in terms of semi empirical method, such as global hardness (η), molecular softness (S), electro philicity index (ω) and electro negativity (χ) are presented in (Table 1). Finally for QSAR/QSPR model building the calculated descriptors set was subjected to two machine learning techniques for the derivation of QSAR models, k Nearest Neighbours (kNN) and symbolic regression by genetic programming (GP).
k-Nearest Neighbors (kNN)
Thek-Nearest Neighbours (kNN) algorithm is based on the idea that the activity of a given compound can be predicted by averaging the activities of its k nearest neighbours, namely, the k compounds most similar to it. This idea follows directly from the similar property principle [29] which states that similar compounds have similar properties. Since chemical similarity between two objects critically depends on molecular descriptors used to characterize them, inherent to kNN is a variable selection procedure which identifies a set of descriptor in terms of which the similar properties principle is satisfied. Furthermore, due to the large number of descriptors subsets, this variable section procedure could be treated as an optimization problem. We have previously implemented the kNN algorithm using Monte Carlo (MC)/Simulated Annealing (SA) as the optimization engine [30] In this work we use our standard set of parameters with 105 steps while replacing a single descriptor and randomly choosing k at each step. The effective temperature was set to produce an initial acceptance rate of ~10% and an average acceptance rate of ~0.5%.
kNN Optimization Based Outlier Removal
Prior to model generation, outliers were removed using a kNN-based outlier removal procedure [30]. Briefly, using kNN, predictions based on short object-neighbours distances are likely to be more accurate than predictions based on long objectneighbours distances. It therefore follows that the activity of an outlier (which is remote from its close neighbours) is likely to be poorly predicted and its removal is likely to improve model performances. Thus, we have devised an algorithm which builds iterative kNN models and removes, at each iteration, that compound whose removal results in the largest improvement in model performances. For outlier removal, we ran the kNN algorithm using the Monte Carlo (MC) optimizer without simulated annealing. The procedure was run for 104 steps per iteration replacing a single descriptor and randomly choosing k at each step. The effective temperature was set to produce an acceptance rate of ~0.5%. The stopping criterion for outlier removal was set to Q2LOO ≥ 0.85.
Library Partitioning
Following outlier removal, the remaining compounds were divided into modelling (75%) and validation (25%) sets. Division was performed with a newly developed method that uses a MC procedure to optimize a representativeness function for the selection of a subset of objects (e.g., compounds) which best represents the parent data set [31]. The unselected and selected subsets constitute the modelling (i.e., training) and validation (i.e., test) sets, respectively. Subsets selected by this method were previously shown to be useful for the evaluation of QSAR models in terms of their ability to predict the activities of compounds residing within their applicability domain [31].
Genetic Programming Based Symbolic Regression for Model Generation
Genetic programming [32] iteratively produce a population of models so that each population contains models that are better than those found in the previous population. This is done by producing an initial set of randomly generated models, ranking them according to their performances and applying a set of operators taken from evolution to the highest ranking ones. Repeating this process for many generations produces a “genetic pressure” which results in the derivation of successively better models until the best model is obtained. Symbolic regression [33] searches the space of mathematical expressions to find the model that best fits a given data set. In this work we used the genetic programming based symbolic regression tool of the Eureqa software [34].
Results and Discussion
Recently, Hui Zhen Zhang et al. [12] reported the synthesis and antimicrobial evaluation of some novel benzimidazole type of Fluconazole analogues with the reference drugs named as Chloromycin, Norfloxacin and Fluconazole. In this report we have tried to establish the relationship between experimental as well as computational theoretical calculated data using DFT based descriptors. In drug research it is a very difficult task to correlate the theoretical calculated data with the experimental data and also to identify the probable site of reaction in any selected bioactive molecules using local density functional based descriptors. Molecular reactivity is expected due to locally but not globally. According to the review of earlier research papers based on QSAR studies it was proved that theoretical descriptors are effective in site selectivity are local softness, fukui functions and local philicity index [6]. Here we used global electron seeking parameters using electrophilicity index, for all the 27 derivatives of benzimidazole type of Fluconazole analogs. Using electrophilicity index descriptor we can easily evaluate the electron donating and electron accepting power of the molecules, moreover while comparing the global electrophilicity index together with the fukui function, local softness and local electrophilicity index we can easily decide the mode of reaction as nucleophilic or electrophilic substitution reaction at the reactive site of a molecule.
Looking at (Figure 1), we divided all the derivatives of a benzimidazole in three groups according to the structure of substitution added at different positions. According to the research studies of Hui-Zhen Zhang et al., all the twenty seven newly synthesized compounds were subjected to in vitro antifungal activity against various fungal cells namely Candida albicans, Saccharomyces cerevisiae and A. flavususing two fold serial dilution technique recommended by National Committee for Clinical Laboratory Standards (NCCLS) with a positive control of clinically antimicrobial drugs Chloromycin, Norfloxacin and Fluconazole [35].
Antifungal Activity
According to the research findings reported by Hui-Zhen Zhang et al., as shown in Table 2 all the newly prepared compounds was tested against all the selected fungal strains (C. albicans, S. cerevisiae and A.flavus). All the target tertiary amine type of benzimidazole derivatives displayed better activities against Fluconazole-insensitive A. flavus. Compound number 4c and 5m exhibits comparable activity against S. cerevisiae as compare to Fluconazole. This showed that newly prepared compounds were less sensitive towards other fungi as compare to reference drug. Compound number 4c showed better activity in compare to other alkyl derivatives having shorter or longer chain length. It was assume that the length of aliphatic chain is mainly responsible for the best antifungal activity. Hence it was observed that (CH2)8 was the most suitable length of alkyl chain for antifungal activity. Compound number 5m named as Bis(trifluoromethyl) phenyl derivative exhibits the highest antifungal activity (MIC = 16-32μg/mL) among the other prepared tertiary amine type of benzimidazole compounds. Compound number 5m exerted better bioactivity (MIC = 16 μg/mL) against C. albicans, S. cerevisiae and A. flavus fungi. Another compound named 3i exhibits strong bioactivity against S. cerevisiae strains (MIC = 2 μg/mL), which is 2- and 8- fold more potent than the Fluconazole which was selected as reference drug respectively [12].
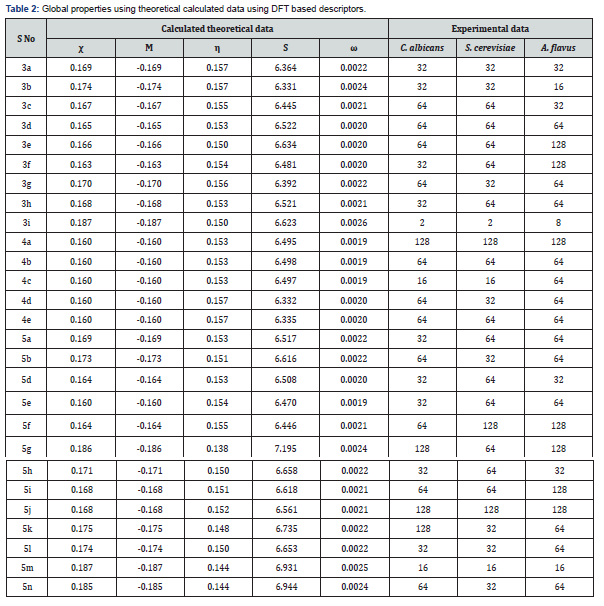
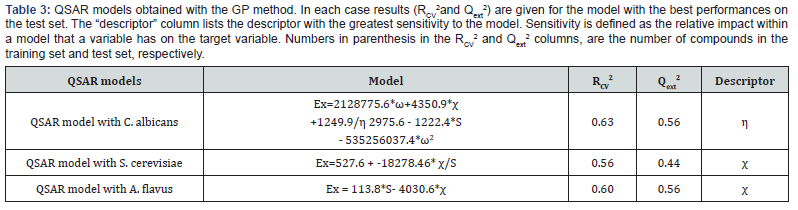
While evaluating the experimental activity data with the calculated theoretical data using DFT based descriptors it shows the same results against the selected fungal strains. Looking at (Table 1), we see that, all the local reactivity descriptors for predicting the probable site of nucleophilic reaction at N3 position of all the selected compounds. When we compared the fukui functions (f+ and f-) values of all the selected compounds then f+ values which is representing the LUMO values are higher than the f- values. Relying upon the relative magnitudes of fukui functions and local softness and local philicity values, we may predict that if the selected compounds undergoes the nucleophilic reaction, then the preferred attacking site is on the N3 position by a molecule. QSAR models were derived using our previously reported workflow [31-36]. A dataset of 27 structures with known antifungal activities in MIC (μM) was collected from the published research paper. The data were manually curated and subjected to descriptors calculation as discussed in the method section. Global descriptors were calculated and following a preprocessing stage, the resulting data set were subjected to an outlier removal procedure [30a]. This procedure led to the no removal of compounds from the data set. The total number of compounds was divided into modelling sets (75%) and test sets (25%), it results into 20 compounds in training and 7 compounds each in test sets for all the three cases (C. albicans, S. cerevisiae and A. flavus) (Table 3) provides the results for models derived by the GP algorithm. A genetic programming approach provides a population of models rather than a single model. Here we present, QSAR models produced by using genetic programming method.
Looking at the (Table 3)values for antifungal activities against C.albians, S.cerevisiae and A. flavus are 0.63, 0.56 and 0.60, respectively. Accordingly, the performances of these models on test sets () are 0.56, 0.44 and 0.56, respectively. Overall, QSAR models with good prediction statistics were obtained with the GP method. The latter method however provides models with varying performances. In conclusion, here we present the QSAR study for the benzimidazole compounds with known MIC (μM). Benzimidazole is an important target for the treatment of various antimicrobial activity. We demonstrate that QSAR models with good prediction statistics could be developed. These models could therefore be used for the design of new benzimidazole compounds [37].
References
- Ansari KF, Lal C (2009) Synthesis, physicochemical properties and antimicrobial activity of some new benzimidazole derivatives. Eur J Med Chem 44(10): 4028-4033.
- Kazimierczuk Z (2002) Synthesis and anti protozoal activity of some 2-(trifluoromethyl)-1H-benzimidazole bioisosteres Acta Biochem Pol 49: 185-195.
- Tominaga T (2001) Jpn. Kokai Tokkyo JP Chem. Abstr.134 93136.
- Grocer H (2002) Synthesis and Anti-fungal Properties of Some Benzimidazole Derivatives, Bioorg Med Chem 10: 2589-2596.
- Ji HT (2003) J Med Chem 46: 474-485.
- Chattaraj PK (2003) J Phys Chem A 107: 4973.
- Parr RG Yang W (1989) Density functional theory of atoms and molecules. Oxford University Press: New York, UK.
- Chatterje A (2003) J Mol Graph Model 25: 208.
- Chattaraj PK, Roy DR (2007) Chem Rev 107: PR46.
- Nakatsuka M, Shimamura T (2001) Jpn Kokai Tokkyo JP Chem Abstr B 170632.
- Shibahara F (2009) 65: 5062.
- Zhang HZ (2013) Design, synthesis and antimicrobial evaluation of novel benzimidazole type of Fluconazole analogues and their synergestic effects with Chloromycin, Norfloxacin and Fluconazole. Eur J Med Chem 64: 329-344.
- Karelson M (2000) Molecular Descriptors in QSAR/QSPR. Wiley Inter science, New York, UK.
- Parr RG (1999) J Am Chem Soc 121: 1922.
- Parr RG, Pearson RG (1983) J Am Chem Soc 105: 7512.
- Yang W, Parr RG (1985) Proc Natl Acad Sci USA 82:6723.
- Chatterje A (2006) J Mol Graph Model 25: 208.
- Fukui K (1982) Science 218: 747.
- Fukui K (1973) Theory of Orientation and Stereoselection. Springer Verlag, Berlin, Germany.
- Parr RG, Yang W (1984) J Am Chem Soc 106: 4049.
- Nakamura H, Yamamoto H (2005) PCT Int Appl Chem Abstr 142: 440277.
- Kim D (2005) A Bioorg Med Chem Lett 15: 2129.
- Wu D (2011) Isolation, structure determination and antibacterial activities of succinamie conjugate diacid from Acinetobacter sp BJ L Microbiol Res 66: 155-160.
- Earl JC, Mackney A (1935) J Chem Soc 53: 899.
- Price S (2009) A Chem Abstr 151: 123970.
- Roy S (2011) Inorg Chim Acta 370: 247
- Shibahara F (2009) J Org Chem 74: 3566.
- Price S (2009) A Chem Abstr 151: 123970.
- Johnson MA, Maggiora GM (1990) Concepts and applications of molecular similarity. John Wiley & Sons, New York, UK.
- Yosipof A, Senderowitz H (2015) J Comput Chem 36: 493-506.
- Zheng W, Tropsha A (1999) J Chemi Inf & Comp Sci 40: 185-194.
- Yosipof A, Senderowitz H (2014) J Chemi Inf & Mod 54: 1567-1577.
- Banzhaf W (1998) Genetic programming: an introduction, Morgan Kaufmann, San Francisco, USA.
- Augusto DA, Barbosa HJ (2000) In Neural Networks Proceedings Sixth Brazilian Symposium, IEEE, Rio de Janeiro, pp. 173-178.
- Schmidt M, Lipson H (2009) Science 324: 81-85.
- (2002) National Committee for clinical Laboratory Standard Document, M27-A2, Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, National Committee for Clinical Laboratory Standards, Wayne, Pennsylvania, USA.
- Yosipof A (2015) Mol Informat 34: 367-379.






























