Ethyl Methane Sulfonate Chemical Mutagenesis of Ba- cillus subtilis for Enhanced Production of Protease
Asia Latif*, Mohsin Iqbal, Muhammad Asgher
Industrial Biotechnology Lab, Department of Biochemistry, University of Agriculture, Pakistan
Submission: February 07, 2018; Published: February 23, 2018
*Corresponding author: Asia Latif and Mohsin Iqbal, Industrial Biotechnology Lab, Department of Biochemistry, University of Agriculture, Faisalabad, Pakistan; Tel: 0313-6763676, E-mail: mohsiniqbal5050@gmail.com
How to cite this article: Asia L, Mohsin I, Muhammad A. Ethyl Methane Sulfonate Chemical Mutagenesis of Bacillus subtilis for Enhanced Production of Protease. Organic & Medicinal Chem IJ. 2018; 5(3): 555664. DOI: 10.19080/OMCIJ.2018.05.555664
Abstract
The enzymes have acquired a great importance in the industries. They are important industrial catalyst used in food, detergent and leather tanning industries. Chemical mutagenesis of Bacillus subtilis was carried out using ethyl methane sulfonate. Mutants of Bacillus subtilis was isolated and evaluated for production of protease to select the best producer mutant strain. Protease production by the best selected mutant was optimized by varying temperature, pH, inoculum size and incubation time in Response Surface Method under Central Composite Design. The mathematical response model is reliable with an R2 value of 0.9833. The adjusted R2 value was 0.9677 suggesting a significant model by determining the close relationship to the actual R2 value. Predicted R2 value shown in this model was 0.9355. The "Pred R-Squared” of 0.9355 is close as to the 'Adj R-Squared” of 0.9677 as expected. The ratio of 31.34 attained in this model represents an adequate signal. The calculated C.V was 2.49 which indicates the good level of model precision and reliability. The maximum enzyme activity was 256.004 (IU/mL) at optimum conditions pH 10, Temperature 35oC, Inoculum size 4mL and fermentation time 120hr. These characteristics render its potential use in detergent industries for detergent formulation.
Keywords: Media optimization; Bacillus subtilis; Chemical mutagenesis; Alkaline detergent proteases; Low-cost media
Abbreviations: RSM: Response surface methodology; PMSF: Phenyl methyl sulphonyl fluoride; DFP: Diisopropyl fluorophosphates; EDTA: Metal chelating agent; UV: Ultraviolet; EMS: Ethyl methane Sulfonate; LB: Luria-Bertania; CCD: Central Composite Design; ANOVA: Analysis of variance
Introduction
Microbial enzymes have been used since ancient Greece in various processes such as cheese making, alcohol production, brewing and baking. Microbial proteases have been used in various industries such as pharmaceutical, food, textile, leather and feed industries. 4.2 billion$ is the present estimate sales of microbial enzymes. Protease enzymes belong to three most important industrial enzymes and their estimated value is $2.21 billion with 6% CAGR from 2016-2021. Microbial Proteases have largest share in industrial markets compare to animal proteases [1]. Among various hydrolytic enzymes microbial proteases are most important enzymes. They are present in animals, plants, fungi and prokaryotes. By fermentation methods in very short time they are cultured in large quantities and produce regular and abundant supply of our desired product. Alkaline proteases have been extensively used as industrial catalysts. In 1914 proteases were introduced as detergent additives and have been used for formulation of detergents [2].
The enzymes have many advantages over conventional chemical method. Enzymes have substrate specificity and high catalytic activity. In worldwide enzyme market, Microbial proteases dominate and having two-third share in detergent industry. Although proteases are produced in all organisms but those microorganisms have been used commercially that produce a considerable number of extracellular proteases [3,4]. Bacillus sp. produced alkaline proteases. The protease has biochemical diversity and widely used in food and tannery industries, detergents and waste treatment, resolution of mixtures of amino acid, medicinal formulations and silver recovery [5]. Bacillus subtilis is best for production of protease [6]. Chemically and UV mutated Bacillus pumilis and Bacillus licheniform is enhance protease production [7]. Thermostable Bacillus amyloliquefaciens also shows protease activity [8]. Protease enzyme is also produced from Bacillus cereus isolated from oyster in marine environment oyster is rich source to produce proteases [9].
PMSF (phenyl methyl sulphonyl fluoride) and DFP (diisopropyl fluorophosphates) completely inhibited protease enzyme. Serine residue present on active site of enzyme sulfonates in presence of PMSF and activity of enzyme is lost. This inhibition profile indicates that these alkaline proteases are serine in nature. Some alkaline proteases are sensitive to EDTA (metal chelating agent) indicating that these proteases are metal ion dependent [10]. Today mutations are very important concept. Mutations cause variations in genes. Mutation is a tool to increase efficiency of microorganisms for production of alkaline protease. Mutant strains enhance production of proteases and lipases [11]. Any heritable change that occurs in DNA sequences and DNA replication is called mutation. Due to this mutation phenotype may be changed [12]. Ultraviolet (UV) light and ethyl methane sulfonate (EMS) are two commonly used mutagens. Mutagenesis increase mutation frequency up to 100-fold/gene without substantial frequency of double mutant and without unnecessary killing of cells. UV and EMS produce different mutant spectra. Mutagenesis by only one type produced significant mutants for study [13]. Mutations that occur under influence of artificial factor are known as induced mutations. Agents that are responsible for mutation are known as mutagens. The chemicals such as ethyl methane sulfonate, methyl methane sulfonate and Ethidium bromide cause mutation and they are strong mutagenic agents. They cause change in DNA replication [14,15]. Chemical mutagens increase mutation frequency and easier to handle. These mutagens can change phenotypic due to deletions, insertions and point mutations in genomic strands [16,17]. EMS is monofunctional alkylating agent. This chemical mutagenic used for DNA lesions. It induces G/C-to-A/T transitions. By SN1/SN2 reaction EMS causes alkylation at N7or O6 of guanine and replaces cytosine with thymine base pair [18]. EMS gives high gene mutations frequency and low chromosome aberrations frequency. Enzyme cost is critical factor that limit use of protease enzyme in various industrial applications. Optimization is defined as a method to enhanced production of protease in classical methods medium conditions optimized by changing single variable optimization strategy. This method has many disadvantages. It is time consuming, it requires more experimental data and interactions among variables interaction is missing [19]. Due to disadvantages, classical method has replaced by statistical optimization like response surface methodology (RSM). To seek optimum conditions in multivariable system RSM is efficient experimental strategy [20].
Experimental

Bacterial Strain: Loop full cells of Bacillus subtilis were transferred to nutrient agar slants and incubated at 370C for 48 hours. The slants were stored in refrigerator (40C). Based on biochemical and morphological characteristics, the colonies were identified. The colonies were subculture occasionally to the fresh slants (Figure 1) [21].
Inoculum Preparation: Inoculum was prepared by adding Luria-Bertania (LB) medium (15.5 g/L) in 250 ml Erlenmeyer flask. The pH (7.5) was adjusted using HCL/NaOH. The medium was autoclaved for 15 minutes at 1210C. After sterilization Bacillus subtilis was inoculated into the flask and shaken at 200 rpm for 48 h at 37°C [22].
Chemical Mutagenesis of Bacillus subtilis by EMS Treatment: Mutation is a random event at the single cell level. One of the most crucial requirements for mutation induction is the selection of an approximate dose of mutagenic agent for mutating the biological materials. The dose can be defined as the dose of a particular mutagen for the definite period of time at the particular temperature. Different EMS concentration of B. subtilis 25μg/ml, 50μg/ml, 75μg/ml and 100μg/ml were used to the bacterial cells inoculation from the solution 1mL was taken and added into 9 mL of medium having fresh bacterial spores. After different time intervals (30, 60, 90, 120 min) EMS treated spores were washed with saline solution (0.15% yeast, 0.89% NaCl) and centrifuged for 10 minutes at 12000 rpm. This step repeated 3 times so that to completely remove mutagen traces. On nutrient agar petri plates washed cells were spread and then incubated for 24 h at 370C.
Liquid State Fermentation for Protease Production: The bacterium was grow in the medium having the following concentration (g/L) Casein 10.0; Maltose, 4.0; Peptone, 1.0; KH2PO4, 0.5; 1.0 Na2CO3; MgSO4. 7H20; 0.02; NaNO3, 0.5 in distilled water. The medium was autoclaved for 15 mints at 121oC and inoculated with B. subtilis. The medium was incubated for 24 hours at 37oC. After 3 days, the broth was centrifuged at 4oC for 20 mints at rotating speed of 10,000rpm and supernatant was used for protease assay [23] (Figure 2).

Analytical Procedures
The method was validated according to ICH guidelines to study system suitability, linearity, precision, accuracy and robustness, limit of detection (LOD) and limit of quantitation (LOQ).
Enzyme Assay: Activity of Protease enzyme was measured by adding casein in phosphate buffer at 37oC. Casein was determined at 660nm using spectrophotometer and by the following equation. Enzyme activity was expressed in U/mL [21].
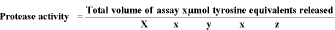
X=volume of enzyme used
Y=Time required for assay (20 mint)
Z=Volume used in colorimetric determination (1ml)
Optimization of Physical Factors for Hyper Production of Protease by the Selected Mutant
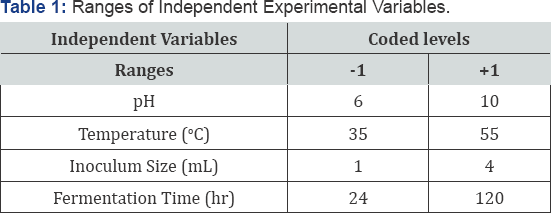
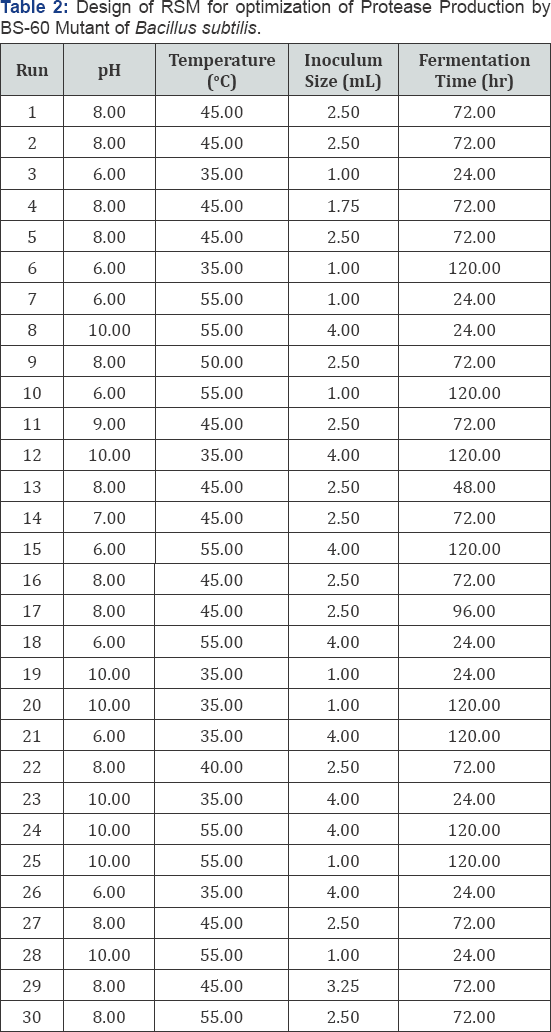
After production of protease by different EMS treated mutants, the best mutant (BS-60 min) was selected. Various parameters effecting the production of protease enzyme included (1) temperature (35-550C); (2) Fermentation period (24-120h); (3) initial pH (6.0-10.0) and (4) inoculum size (1-4mL) using Response Surface Methodoly (RSM) under Central Composite Design (CCD) by using design expert version 7.1 (Table 1). Varying different factors simultaneously their interactive effect on responses was analyzed by RSM. The Central Composite Design (CCD) was used to study interaction effects of various factors on protease enzyme production. For this research work 4 factors design has been employed in second order polynomial model that indicate 30 runs were required (Table 2). In 250 ml flasks of different pH media were prepared and autoclaved. Substrate was inoculated by varying B. subtilis inoculum size in each flask. The contents were mixed and incubated for different incubation time at different temperatures.
Results and Discussion
Bacillus subtilis IBL-04 was used for chemical mutagenesis to produce protease in LSF using casein as substrate. Optimum growth conditions like pH, temperature, inoculums size and incubation time were also determined for hyper production of protease from selected mutant Bacillus subtilis IBL-04.
Bacterial Strain Improvement for Hyper Production of Protease
Productivity of enzymes can be enhanced by Strain improvement techniques followed by process of optimization. Mutagenesis is cost effective and easiest source which create mutations in living system. Physical or chemical mutagenesis is done for hyper production of industrially important strain. In double mutations both chemical and physical mutagenesis may be applied treating wild type strain with UV-radiations followed by chemical mutagenesis [24].
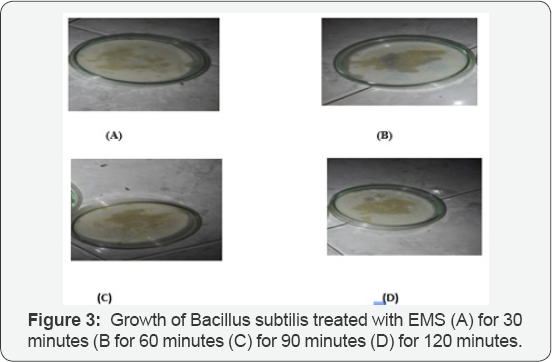
Mutagenesis by EMS Treatment: Mutation in microorganisms mainly depends on type of mutagen, dose of mutagen and time of exposure. An optimum EMS mutagen dose of 100μg/mL was applied and bacterial cell suspension was treated for different time period 30, 60, 90 and 120 minutes. Treatment time of 60 min (EMS-60) gave the best results and bigger sized colonies on petri plates. EMS-60 was therefore, selected as best mutant as shown in Figure 3.
Selection and Evaluation of Mutants
After mutagenesis spores of B. subtilis were 6-fold diluted by pouring 0.1 ml of mutagen from dilutions on skim milk agar plates. EMS-60 gave bigger size colonies on petri plates. For selection of mutagenic strains, 2-Deoxy-D-glucose (1%) was used which is non metabolizable and toxic analogue of glucose. It has potential to restrict the growth of parent strain but mutant strain can grow on 2-Deoxy-D-glucose containing media (Figure 4). The mutants growing on selection medium was picked and transferred to growth restriction medium.
Colony restriction using triton X-100
For the selection of colonies 2% triton x 100 was used in selection media. Triton x100 was effective colony restrictor and it gave good round colonies on skim milk agar plates (Figure 5). Triton X-100 is denaturing, stabilizing and nonionic detergent. It can reduce the bacterial and fungal growth by delaying the growth of microbes.
Production of protease by selected mutants
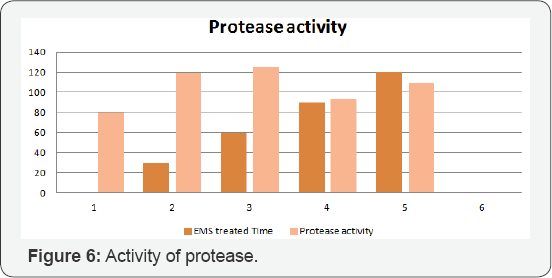
Mutated strains BS-30, BS-60, BS-90 and BS-120 from EMS mutagenesis of Bacillus subtilis IBL-04 were used for protease production in liquid state fermentation medium of casein. The uniform conditions of fermentation media (pH 7.5, 2ml inoculum) were applied at 370C for 24 hours incubation. BS- 60 was selected as best protease producer as it yielded highe protease activity with as compared to parent and then mutant strains. Results showed that mutagenesis of Bacillus subtilis enhanced the protease productivity. The mutated strains showed higher enzyme production as compare to \normal parent strain (Table 3) (Figure 6).

Optimization of fermentation parameters by RSM under CCD
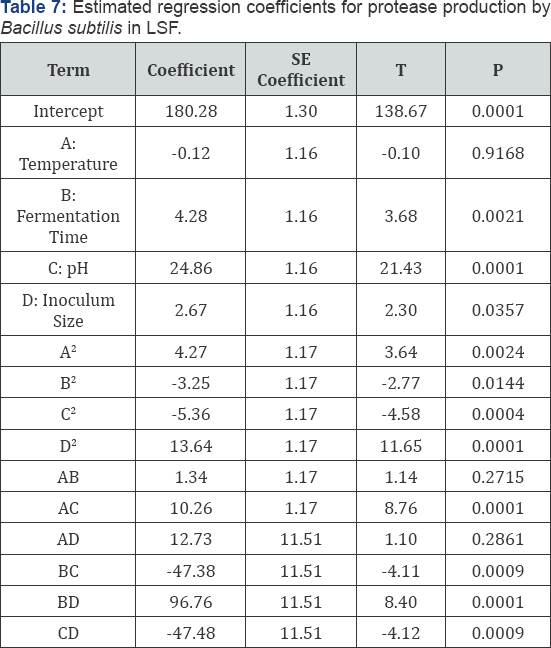
Foe hyper production of protease by selected mutant BS- 60, various parameters like pH, temperature, inoculum size and fermentation time were optimized using casein as substrate in LSF. A mathematical and statistical technique was invented by Wilson and Box which is known as Response Surface Methodology (RSM). RSM was used to optimize the variables simultaneously. ANOVA (Analysis of variance) was used to calculate predicted response (Protease activity) and identify significant factors. Using RSM independent variables Temperature (A), Fermentation Time (B), pH (C) and inoculum size (D) and their interactional effects were investigated on protease production by BS-60 mutant. Triplicate flasks were processed and optimum values were placed in low order polynomial equation to find best response. In 30 runs design four factors were studied. After carrying experiments protease activity (Responses) were measured. Response values were obtained as mean of triplicate runs. The optimal protease activity was shown at 4 mL inoculum size, 120 h fermentation time, pH 10 and temperature 350C.
Protease assay using Casein (substrate) was employing and Enzyme (protease) activity was measured at 660nm. Protease yield was expressed in U/mL. Highest protease activity was 256.004U/mL while minimum 153.013 U/mL using same substrate casein (Table 4) RSM has been used for optimization of fermentation processes, enzymatic processes and catalytic studies. For different coefficients of model, CCD gives the most appropriate and precise estimates and reduce ellipsoidal confidence level. Response surface methodology contain group of techniques analyzed. The relations between measured responses and controlled experimental factors according selected criteria. Test factors had been coded to develop regression equation:
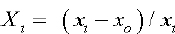
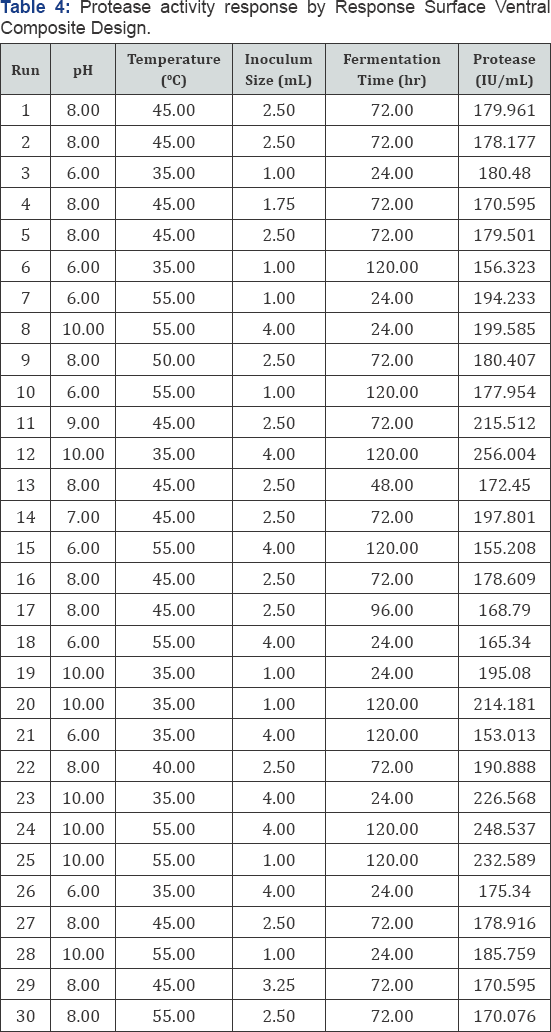
To study each variable effect and their interaction effect following equation used
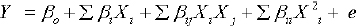
Y is measured (predicted) response, pi,, pij and pii are linear, square and quadratic coefficients while e is random error [25].
Final Equation in Terms of Coded Factors:
This equation is used to predict response of each factors along with their sublevels. high level of factors coded as +1 and low levels as -1.
ANOVA for the Response (protease) Surface Model
The "Pred R-Squared” of 0.9355 is in reasonable agreement with the "Adj R-Squared” of 0.9677. Determination coefficient (R2) was used to check aptness of model. In this study, the value of coefficient of determination (R2=0.9833) shows that 1.67% of the total variations was not explained by the model. The value of Adj. R2=0.9677 is a very high and indicates a high significance of the model. A higher value of the correlation coefficient (R2 = 0.9355) signifies an excellent correlation between the independent variables (18). At the same time, a relatively lower value of the coefficient of variation (CV=2.49) indicates improved precision and reliability of the conducted experiments (Table 5).

The results of analysis of variance (ANOVA) are shown in the Table 6. The F test and subsequent P values with the factors were predicted. The lesser the P value, the greater is the importance of corresponding coefficient. The Model F-value of 63.11 implies the model is significant. There is only a 0.01% chance that a "Model F-Value” this large could occur due to noise.Values of "Prob > F” less than 0.0500 indicate model terms are significant. In this case B, C, D, AB, AC, AD, BC, CD, B2, C2, D2 are significant model terms. Values greater than 0.1000 indicate the model terms are not significant. The "Lack of Fit F-value” of 1.90 implies the Lack of Fit is not significant relative to the pure error. In this case there is a 24.82% chance that "Lack of Fit F-value” due to noise (Table 6).
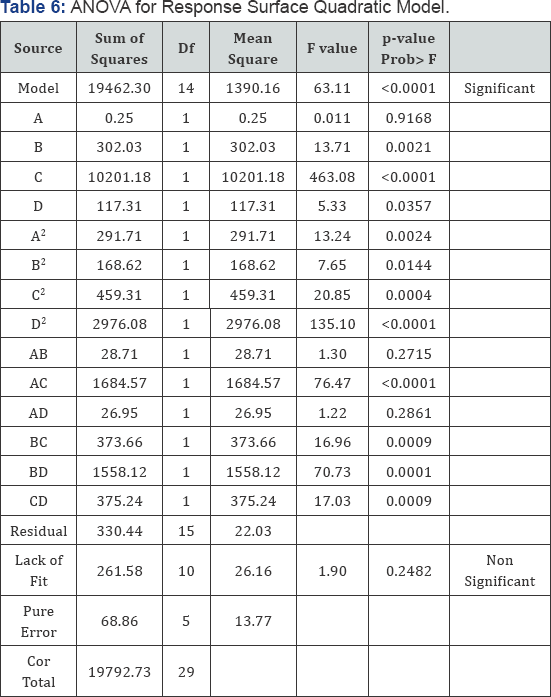
Regression Coefficient for Protease enzyme production from Bacillus subtilis
In case of regression coefficient model, the positive and negative coefficient of any factor indicates that it has significant effect on protease enzyme production. The positive values of linear coefficient indicates that production of protease enzyme increased with initial increase in any factor. While the negative linear coefficient for pH, temperature, inoculum size and fermentation time indicated that at higher levels of these factors protease activity decreased. The t and p values are usually applied to analyze the significance of each coefficient. Greater t test value and lower the p value depicts greater significance of corresponding coefficient. P value P≤0.01 predicts that model terms are highly significant (Table 7).
Graphical Representation of Interaction among Variables
Response surface graphs have been used to elucidate the interactive effect of different variables and get the optimal point of all the variables to obtain maximum yield. For optimum production of protease, optimal value of the each variable and interaction between various physicochemical parameters have been explained by constructing (3D) graphs. 3D response graphs show the optimum leve of response and interaction of variables.
Temperature vs Fermentation Time: Response Surface Plot exhibited the high protease activity synthesized by Bacillus subtilis at 350C temperature and 120 hours fermentation time. The Response Surface Plot predicted the highest protease activity of 256.004 IU/mL. There was a significant influence of the interaction of temperature and fermentation time on protease production by BS-60 mutant (Figure 7).
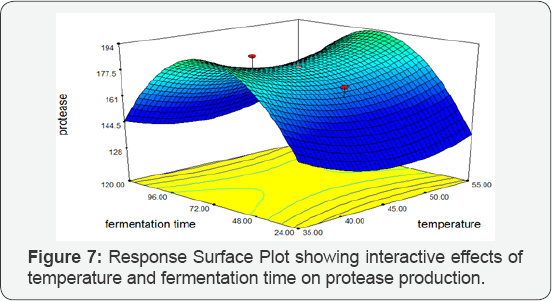
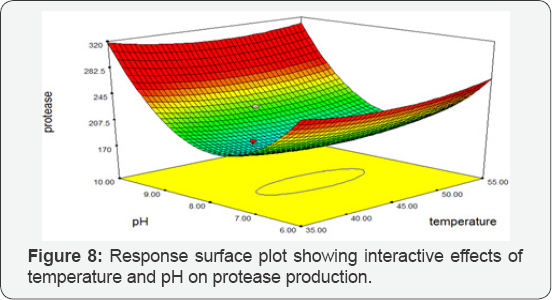
Temperature vs pH: Response Surface Plot exhibiting the high protease activity synthesized by Bacillus subtilis at 350C temperature and pH 10. The Response Surface Plot predicted the greatest protease activity of 256.004 IU/mL. There was a significant influence of the interaction of temperature and pH on protease production by BS-60 mutant (Figure 8).
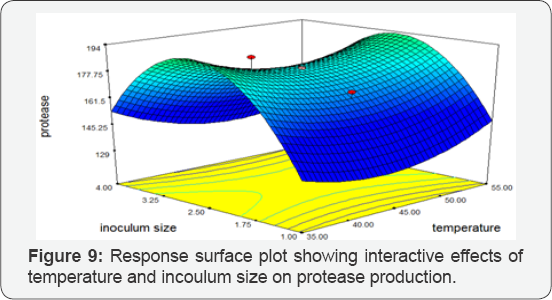
Temperature vs Inoculum Size: Response Surface Plots exhibiting the high protease activity synthesized by Bacillus subtilis at 350C temperature and 4mL. The 3D plot predicted the greatest protease activity of 256.004 IU/mL. There was a significant influence of the interaction of temperature and inoculum size on protease production by BS-60 mutant (Figure 9).
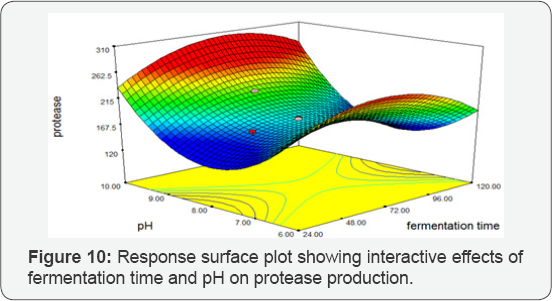
Fermentation Time vs pH: Response Surface Plots exhibiting the high protease activity synthesized by Bacillus subtilis at 120 hrs and10 pH. The 3D plot predicted the greatest protease activity of 256.004 IU/mL. There was a significant influence of the interaction of fermentation time and pH on protease production by BS-60 mutant (Figure 10).
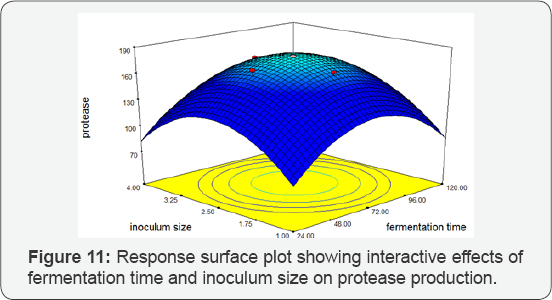
Fermentation Time vs Inoculum Size: Interaction effect of inoculum size and fermentation time was significant. Protease gives optimum activity at 120 hrs fermentation time and 4mL inoculum size. The 3D plot predicted the greatest protease activity of 256.004 IU/mL. There was a significant influence of the interaction of fermentation time and inoculum size on protease production by BS-60 mutant (Figure 11).
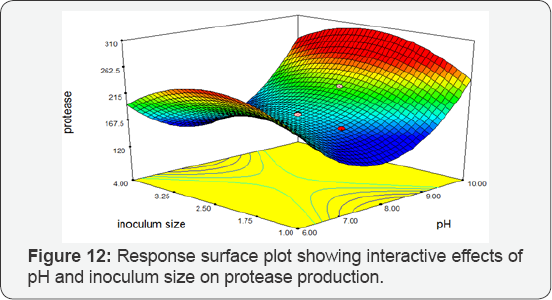
pH vs Inoculum Size: Response Surface Plot exhibiting the high protease activity synthesized by Bacillus subtilis at pH 10 and 4 mL inoculum size. The Response Surface Plot predicted the greatest protease activity of 256.004 IU/mL. There was a significant influence of the interaction of pH and inoculum size on protease production by BS-60 mutant (Figure 12). It was reported that the constant and highest protease enzyme activity was observed at pH between 6-10°C. The optimum pH for alkaline protease was observed at 10°C. The production at pH 9 and 10 are mostly comparable [26]. The maximum enzyme activity was 256.004 (IU/mL) at optimum pH 10. It means pH 10 is more favorable for protease production. It was also observed that if inoculum size was reduced (0.2%) so it may cause less protease enzyme production. Pseudomonas sp. has been reported that 1.5% inoculum showed maximum enzyme production [27]. Similarly, Bacillus subtilis has 4% inoculum size which showed maximum enzyme production. It is relatively comparable. Halobacterium sp. has been reported that maximum protease production was achieved 96 h incubation period [28]. The Bacillus subtilis gave maximum protease production in 120 h incubation time.
Conclusion
The results obtained confirmed that Chemical mutagenesis technique is an important tool in Bacillus improvement for increasing production of alkaline protease enzyme. Thus, the selected Bacillus mutant has potential in biotechnological applications.
References
- Singh R, M Kumar, A Mittal, PK Mehta (2016) Microbial enzymes: industrial progress in 21st century. 3 Biotech 6(2): 174.
- Gupta R, Q Beg, P Lorenz (2002) Bacterial alkaline proteases: molecular approaches and industrial applications. Appl Microbiol Biotechnol 59(1): 15-32.
- Arunkumar T, A Anand, G Narendrakumar (2014) Application of response surface methodology (RSM)-CCD for the production of laccases using submerged fermentation. International Journal of Pharma and Bio Sciences 5(4): 429-438.
- Mohsin I, Muhammad A, Fareeha B (2017) Development of Bacillus subtilis Mutants for Overproduction of Protease. J Microb Biochem Technol 9(4): 174-180.
- Agrawal R, R Singh, A Verma, P Panwar, AK Verma (2012) Partial purification and characterization of alkaline protease from Bacillus sp. Isolated from soil. World J Agric Sci 81: 29-133.
- Geethanjali S, S Anitha (2011) Optimization of protease production by Bacillus subtilis isolated from Mid Gut of Fresh Water Fish Labeo rohita. J Fish and Marine Sci 3(1): 88-95.
- Nadeem M, Q Syed, A Liaquat, S Baig, A Kashmiri (2010) Study on biosynthesis of alkaline protease by mutagenized culture of Bacillus pumilus. Pak J Food Sci 20: 24-30.
- Mrunmaya KP, KS Mahesh, T Kumananda (2013) Isolation and characterization of an Odisha, India. Int J Microbiol 5:159-165.
- Umayaparvathi S, Meenakshi S, Arumugam M, Balasubramanian T (2013) Purification and characterization of protease from Bacillus cereus SU12 isolated from oyster Saccostrea cucullata. African Journal of Biotechnology 12(40): 5897-5908.
- Debananda SN, K Pintubala (2010) A Thermostable Alkaline Protease from a Moderately Halo-alkali thermotolerant Bacillus subtilis Strain SH1. J Basic Appl Sci 4: 5126-5134.
- Soliman EAM, WK Hegazy, EM Maysa (2004) Induction of overproducing alkaline protease Bacillus mutants through UV irradiation. Arab J Biotech 8: 49-60.
- Ralph, Kirby (2003) Fundamentals of biochemistry, cell biology and biophysics. Vol. II, Prokaryote Genetics and small effects. Science 317: 813-815.
- Saxena S (2015) Strategies of Strain Improvement of Industrial Microbes. Appl Microbiol 155-171.
- Haq IU, H Mukhtar, H Umber (2006) Production of protease by Penicillium chrysogenum through optimization of environmental conditions. J Agric Social Sci 2(1): 23-25.
- Besoain XA, LM Perez, A Araya, L Lefever, JR Montealegre (2007) New strains obtained after UV treatment and protoplast fusion of native Trichoderma harzianum: their biocontrol activity on Pyrenochaeta lycopersici. E J Biotechnol 10(4): 604-617.
- Greene EA, CA Codomo, NE Taylor, JG Henikoff, BJ Till, et al. (2003) Spectrum of chemically induced mutations from a large-scale reverse- genetic screen in Arabidopsis. Genetics 164(2): 731-740.
- Flibotte S, ML Edgley, I Chaudhry, J Taylor, SE Neil (2010) Whole- genome sequencing profiling of mutagenesis in Caenorhabditis elegans. Genetics 185(2): 431-441.
- Sikora P, A Chawade, M Larsson, J Olsson, O Olsson (2012) Mutagenesis as a tool in plant genetics, functional genomics, and breeding. Int J plantgenomics.
- Li X, T Xu, X Ma, K Guo, L Kai, et al. (2008) Optimization of culture conditions for production of cis-epoxysuccinic acid hydrolase using response surface methodology. Bioresour Technol 99: 5391-5396.
- Montgomeryd DC, GC Runger (2002) Applied Statistics and Probability for Engineers (3rd edn.); John Wiley and Sons, New York, USA.
- Pant G, A Prakash, JVP Pavani, S Bera, GVNS Deviram, et al. (2015) Production, optimization and partial purification of protease from Bacillus subtilis. J Taibah University Sci 9(1): 50-55.
- Mukhtar H, IU Haq (2012) Purification and characterization of alkaline protease produced by a mutant strain of Bacillus subtilis. Pak J Bot 44: 1697-1704.
- Mrudula S, Begum AA, Ashwitha K, Pindi PK (2012) Enhanced Production of Alkaline Protease by Bacillus subtilis In Submerged Fermentation. Int J Pharm Bio Sci 3(3): 619-631.
- Xu D, JC Cote (2003) Phylogenetic relationships between Bacillus species and related genera inferred from comparison of 3' end 16S rDNA and 5' end 16S-23S ITS nucleotide sequences. Int J Syst Evol Microbiol 53: 695-704.
- Adinarayana K, P Ellaiah, DS Prasad (2003) Purification and partial characterization of thermostable serine alkaline protease from a newly isolated Bacillus subtilis PE-11. Aaps Pharmscitech 4(4): 440-448.
- Joo HS, CS Chang (2005) Production of protease from a new alkalophilic Bacillus sp. I-312 grown on soybean meal: optimization and some properties. Proc Biochem 40(3-4): 1263-1270.
- JR Dutta, PK Dutta, R Banerjee (2004) Optimization of culture parameters for extracellular protease production from a newly isolated Pseudomonas sp. using response surface and artificial neural network models. Process Biochem 39(12): 2193-2198.
- SV Anand, J Hemapriya, J Selvin, S Kiran (2010) Production and optimization of haloalkaliphilic protease by an extremophile- Halobacterium sp. Js1, isolated from thalassohaline environment. Global J Biotechnol Biochem 5(1): 44-49.






























