QSAR and Molecular Interaction Study of Piperine Analogues for Antitubercular Activity
Sakshi Bhardwaj and Sonal Dubey *
Department of Pharmacy, Krupanidhi College of Pharmacy, India
Submission: July 20, 2017; Published: July 31, 2017
*Corresponding author: Sonal Dubey, Krupanidhi College of Pharmacy, Chikka Bellandur, Carmelaram Post, Karnataka, India; Email: drsonaldubey@gmail.com
How to cite this article: Sakshi B, Sonal D. QSAR and Molecular Interaction Study of Piperine Analogues for Antitubercular Activity. Organic & Medicinal Chem IJ. 2017; 3(2): 555606. DOI: 10.19080/OMCIJ.2017.03.555606.
Abstract
In the present work QSAR and molecular docking studies have been performed to explore the binding affinity of 70 novel piperine analogues and 23 reported compounds against Mycobacterium tuberculosis (strain ATCC 25618 / H37RvJ. Molecular docking studies for training and test compounds were done against protein Dev R fUniprot ID: P9WMF8J. The DevR-DosR works on two component regulatory system and was concerned in dormancy response of Mycobacterium tuberculosis. The best model from the training set showed r2 value 0.8760 and q2 value 0.7516. The validation of best QSAR model of each series was done by predicting the activity of the test set compounds. We have found that the reported compounds interacted with protein with a range of binding energy from -2.31 k cal/mol to -4.941 kcal/molby formation of one hydrogen bond to four hydrogen bond whereas predicted compounds interacted with binding energy ranges from -2.36 kcal/mol to -5.90kcal/ molby forming one hydrogen bond to five hydrogen bonds.Similar pharmacophores containing molecules were designed and their activities were predicted using validated QSAR model and docking scores were also calculated. Some of predicted compounds showed improved binding affinity with the selected protein and some of them showed comparable affinity as compare to reported compounds.
Keywords: QSAR, Tuberculosis, In Silico, Piperine, Verapamil
Abbreviations: QSAR: Quantitative Structure Activity Relationship; TB: Tuberculosis; MDR-TB: Multi Drug Resistant Tuberculosis; XDR-TB: Extensively Drug Resistant Tuberculosis; WHO: World Health Organization; TRPV: Transient Receptor Potential Vanilloid
Introduction
Tuberculosis fTBJ is a bacterial infection caused mainly by Mycobacterium tuberculosis, most commonly affects the lungs. According to WHO report an estimated 1.8 million people died from TB in 2015, of whom 0.4 million were co-infected with HIV. MDR-TB remains a public health crisis. Three countries carry the major burden of MDR-TB -India, China, and the Russian Federation. World Health Organization updated the estimate of incidence in India- that is, the number of new tuberculosis cases in a year - from 1.7 million cases to 2.8 million in 2015. In 2015, there were an estimated 10.4 million new (incident) TB cases worldwide, of which 5.9 million (56%) were among men, 3.5 million (34%) among women and 1.0 million (10%) among children. Six countries accounted for 60% of the new cases: India, Indonesia, China, Nigeria, Pakistan and South Africa. Worldwide, the rate of decline in TB incidence remained at only 1.5% from 2014 to 2015 [1]. There were an estimated 1.4 million TB deaths in 2015, and an additional 0.4 million deaths resulting from TB disease among people living with HIV. There are nine drugs in advanced phases of clinical trials for the treatment of drug- susceptible TB, drug-resistant TB or LTBI. These are bedaquiline, delamanid, linezolid, PBTZ169, pretomanid, Q203, rifampicin (high-dose), rifapentine and sutezolid. There are 13 vaccine candidates in clinical trials, including candidates for prevention of TB infection and candidates for prevention of TB disease in people with LTBI.
The symptoms of active TB of the lung are coughing, sometimes with sputum or blood, chest pains, weakness, weight loss, fever, and night sweats [2]. The chemical composition of the mycobacterium cell wall and its unusual structurecreates major difficulty in TB treatment hence makes many antibiotics ineffective.Paleopathology and paleoepidemiology development in infectious diseases has proven the origin of this disease [3]. In 1993, the World Health Organization (WHO) declared TB to be a global emergency [4]. The exact cause of TB is unknown; it is thought that it could be because of the occurrence of HIV infection as well as MDR-TB due to inefficient management. Isoniazid and Rifampicin are the two main drugs used in current first-line anti-TB chemotherapy. The therapy with existing TB drugs is exceedingly lengthy. Whereas MDR-TB and XDR-TB further complicates the world situation [5].
Poor activity of existing therapies and increasing drug resistance towards the latent stage of Mycobacterium tuberculosis has produced a clear need to develop novel therapeutics [6]. Natural products including plants, animals and minerals have been the basis of treatment of human diseases since from an ancient time. However, many effective medicines, including morphine, ephedrine, reserpine, aspirin, atropine and digitoxin were developed from natural products [7]. In our present work, we have focused on piperine, an alkaloid, major chemical constituent present in piper species. This alkaloid is responsible for pungency of black pepper. The pungency of piperine is because of activation of the heat and acidity sensing Transient receptor potential vanilloid (TRPV) ion channel TRPV1 on nociceptors [8].
Here In silico approach used as a strategy for designing and predicting activities of predicted compounds in comparison with some reported compounds. Verapamil analogues are derivatives of dimethoxy phenyl ring attached to a carbon chain have proven themselves as good anti-tubercular agents whereas piperine also having methylenedioxyphenyl ring attached to a carbon chain which is found to be essential structural requirement for its activity. Hence in present work we correlated these two compounds for same activity. QSAR is a methodology to design a rational molecule that meets the above-said requirement with not much effort, less time and lesser issues of environmental pollution. A QSAR equation correlates variety of physical or chemical parameters with biological activity [9-12]. There are many examples available in literature of successful screening of active compounds by QSAR methodology [13,14]. In our study we predicted QSAR model from training compounds and further used that model for activity prediction of test compounds.
Materials and Methods
A total of 23 dimethoxyphenyl derivatives were selected from literature for QSAR and docking studies which have reported their activity against Mycobacterium tuberculosis. All the structures had the same pharmacophore with variable substitutions are which contributes to the difference in the observed anti tubercular activity.
QSAR studies
Around 800 descriptors for each of the compound were calculated using DRAGON and Chem office softwares. MLRA (Multiple Linear Regression Analysis) analysis was performed on each series to get best QSAR model using CODESSA®. Each series' best model was validated by predicting the activity of all training set compounds. The software enables evaluation of molecular descriptors and builds regression equation relating the best set of descriptors with the activity which can be used later for predicting activity of new molecules. The structures of the compounds were drawn and optimized by Chem Draw software. Eight hundred descriptors (Physico-chemical, Alignment Independent and Atom Type descriptors) were estimated. The data was divided into training and test sets randomly. Multiple linear regression analysis Method was used to identify the best model. Statistical parameters as r2, q2, S2 and F were estimated for the regression equation to determine the quality of the data fit and the predictive capability for the model. Hence, we have designed 70 novel compounds by predicting their activities with the help of QSAR and docking results.
Docking Studies
The software used for studying the drug receptor interaction and designing new molecule was GLIDE module of Schrodinger 2016-1. Protein (3C3W) structure was obtained from www.rcsb. org and the protein was prepared using the module Protein Prep Wizard. The site mapping was done to get the binding cavity within the protein which is further used for the studies. Ligand structures were drawn by using 2D sketcher and optimized. Energy optimization was done using OPLS3. The pH of the simulated environment was maintained between 7±2. Molecular docking studies were done to find out the interactions between reported as well as predicted molecules. XP docking method was used in the GLIDE module to dock the ligands within the binding cavity of protein.
Results and Discussion
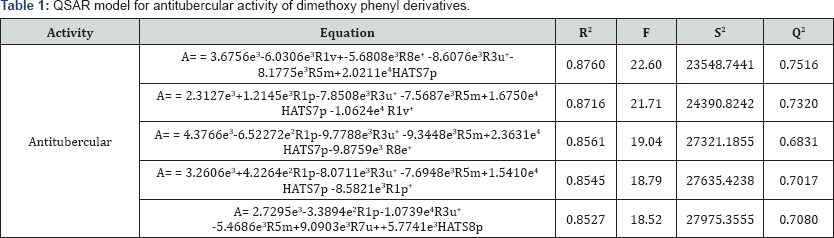

Using 23 analogues bearing dimethoxy phenyl ring QSAR models were generated. The best fit model from QSAR studies for antitubercular activity showed r2 value 0.8760 and q2 value 0.7516, details are given in Table 1. The best equation shows that the anti TB activity is dependent on R1v (vander Waals volume), R8e+ (Sanderson electronegativity), and HATS7p (polarizability). Figure 1 is showing the correlation plot between calculated and experimental activities of antitubercular dimethoxy phenyl derivatives.
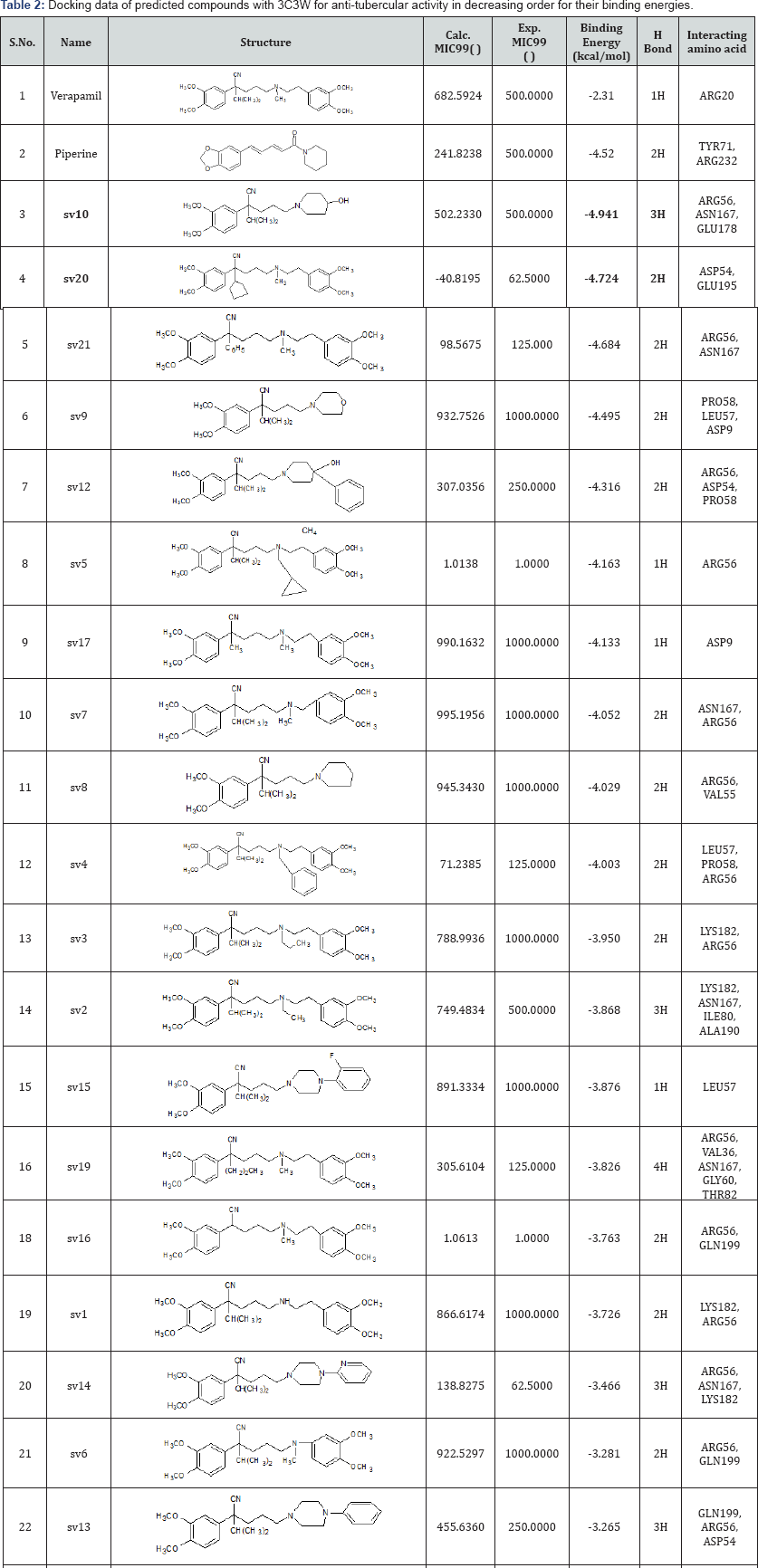
The molecular docking studies results in binding energy, binding affinity and interaction of ligands with the selected protein against Mycobacterium tuberculosis. The calculated and experimental activities, binding energies, hydrogen bond formed and interacting amino acids are given in Table 2 for reported compounds. Whereas calculated activity, binding energy, hydrogen bond formed and interacting amino acids for predicted compounds are given in Table 3. We have found that the reported compounds interacted with protein with a range of binding energy from -2.31 k cal/mol to -4.941 kcal/ molby formation of one hydrogen bond to four hydrogen bond whereas predicted compounds interacted with binding energy ranges from -2.36 kcal/mol to -5.90kcal/mol to by forming one hydrogen bond to five hydrogen bonds. The Figure 2 (a-h) shows the docked molecules sv10, sv20, p622, p1057 and p545 with good binding energy inside the binding cavity of protein. The amino acids majorly interacting with ligand at binding site of protein are GLN199, THR198, ALA200, VAL185, LEU57, PRO58, THR166, LEU165, VAL55, ARG56, LEU161, and GLY60. Schiff's bases were here proven to be good active compounds with better binding affinity against selected protein. In present study, electronegative atoms attached to Schiff's bases are contributing towards desired activity.


Conclusion
Based on QSAR and molecular docking studies performed on 23 reported including reference compounds and 70 predicted compounds for antitubercular activity showed that binding energies for reported compounds with 3C3W protein ranges from -2.31 k cal/mol to-4.941 kcal/mol with one hydrogen bond to four hydrogen bonds. Whereas binding energies for predicted compounds with one hydrogen bond to five hydrogen bonds ranges from -2.36 kcal/mol to -5.90 kcal/mol. Some of the predicted compounds have shown good binding affinity as compared to reference drugs. Thus, we can be concluded that the QSAR models generated are good as the docking scores are comparable these models can be used to predict anti TB activity of the new compounds.
Conflict of Interest
Authors do not hold any economic interest in this work, and they do not hold any conflict of interest in the work presented.
References
- WHO report, Treatment of tuberculosis, 2015-2016.
- 2014 Tuberculosis.
- Godreuil S, Tazi L, Banuls AL (2007) Pulmonary Tuberculosis and Mycobacterium Tuberculosis: Modern Molecular Epidemiology and Perspectives. Encyclopedia of Infectious Diseases: Modern Methodologies 1: 1-29.
- Sharma SK, Mohan A (2004) Multidrug-resistant tuberculosis. Indian J Med Res. 120: 354-376
- World Health Organization (WHO) (2009) Treatment of tuberculosis: Guidelines for national programmes, Geneva: World Health Organization (4th edn), Geneva, Switzerland.
- Sacchettini JC, Rubin EJ, Freundlich JS (2008) Drugs versus bugs: in pursuit of the persistent predator Mycobacterium tuberculosis. Nat Rev Microbiol 6(1): 41-52.
- Kurokawa M, Shimizu T, Watanabe W, Shirak K (2010) Development of New Antiviral Agents from Natural Products. The Open Antimicrobial Agents Journal 2: 49-57.
- McNamara FN, Randall A, Gunthorpe MJ (2005) Effects of piperine, the pungent component of black pepper, at the human vanilloid receptor (TRPV1). Br J Pharmacol 144(6): 781-790.
- Hansch C, Kurup A, GargR, Gao H (2001) Fragment-based QSAR: perspectives in drug design. Bioorg Med Chem 101: 619-672.
- Maloney PP, Hansch C, Fujita T, Muir RM (1962) Computational biology and Quantitative structure-activity relationship. Nature 194: 178-180.
- Fujita T, Iwasa J, Hansch C, Am J (1964) A Method for the Correlation of Biological Activity and Chemical Structure Chem. Soc 86: 5175-5180.
- Hansch C (1969) Quantitative approach to biochemical structure- activity relationships. Acc Chem Res 2(8): 232-239.
- Shi LM, Fan Y, Myers TG, Paull KD, Weinstein JN, et al. (1998) Data Mining: An Integrated Approach for Drug Discovery. J Chem Inf Comput Sci 38: 189-199.
- Oloff S, Mailman RB, Tropsha A (2005) Application of validated QSAR models of D1 dopaminergic antagonists for data-base mining. J Med Chem 48(23): 7322-7332.






























