The Contribution of Long Noncoding RNAs Inpancreatic Adenocarcinoma; in Silico Analysis
Hossein Ansari1,2*, Maryam Tahmasebi Birgani2,3, Arman Shahrisa4, William Cho5 and Mojgan Khodadadi1
1Department of Biotechnology, Ahvaz Branch, Islamic Azad University, Iran
2Department of Medical Genetics, School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Iran
3Cellular and Molecular Research Center, Ahvaz Jundishapur University of Medical Sciences, Iran
4Department of Molecular Genetics, Faculty of Biosciences, Tarbiat Modares University of Tehran, Iran
5Department of Clinical Oncology, Queen Elizabeth Hospital, China
Submission: May 11, 2018; Published: May 31, 2018
*Corresponding author:Hossein Ansari, Department of Biotechnology, Ahvaz Branch, Islamic Azad University, Ahvaz, IR Iran, Tel: +989163001067; Email: hosseinansari62@gmail.com
How to cite this article: Hossein A, Maryam T B, Arman S, William C, Mojgan K. The Contribution of Long Noncoding RNAs Inpancreatic Adenocarcinoma; in Silico Analysis. J Tumor Med Prev. 2018; 3(2): 555606. DOI: 10.19080/JTMP.2018.03.555606
Abstract
Long noncoding RNAs (lncRNAs) are lengthy noncoding transcripts with important roles in regulation variety of cellular and molecular pathways in the cell like cell proliferation, apoptosis and differentiation. Therefore, it is not surprising if their altered expression connected with human diseases especially cancers. Here, we investigated the expression levels of 192 approved lncRNAs in pancreatic adenocarcinoma using available cancer bioportals. Compared to other lncRNAs, the expression levels of lncRNAsGAS5, NORAD, XIST, ZFAS1 and PVT1 were altered in more patients. Additionally, these five genes were upregulated in tumorous tissues with z-score more than 3. These finding indicates that these lncRNAs are promising gene targets for experimental studies on pancreatic adenocarcinoma which may eventually lead to find a novel biomarker for this cancer.
Keywords: Long noncoding RNA; Cancer; Pancreatic adenocarcinoma; GAS5; NORAD; XIST; ZFAS1; PVT1
Introduction
Long noncoding RNAs (lncRNAs) are lengthy transcripts with no capacity for protein production [1]. Increasing body of evidences have been demonstrated that lncRNAs play an important role in regulation of variety of cellular and molecular pathways in the cell including cell growth, proliferation, apoptosis and differentiation [2,3]. Therefore, it is not unexpected if their aberrant expression was attributed with human tumors. The contribution of lncRNAs in progression and invasion of human cancers including breast, gastric, hepatocellular carcinoma and colorectal adenocarcinoma [4] was subject of variety of studies. Meanwhile, valuable feature of lncRNAs like their tissue-specific expression and the long-term stability in body fluids like serum, urine, saliva, breast milk and cerebrospinal fluids, has attracted the cancer researchers to focus more and more on these noncoding molecules as novel marker of human cancers [5]. Pancreatic adenocarcinoma is the fourth cause of cancer- related death worldwide and can progress due to the presence of some risk factors like cigarette smoking, obesity or exposure to chemicals like beta-naphthylamine and benzidine [6]. As the pancreas plays vital role in production of digestive enzymes and hormone, diagnosis and treatment of pancreatic lesions is essential [7]. The involvement of lncRNAs in pancreatic adenocarcinoma is recently proposed by some studies, however, the need for further experiments is felt [8]. Owing to this fact, this study was aimed to analyze the expression levels of all approved lncRNAs in pancreatic adenocarcinoma tissues using available cancer bioportal. This study proposes which lncRNAs are the best candidates for experimental researches on pancreatic adenocarcinoma.
Material and Methods
To find the association of lncRNAs with pancreatic adenocarcinoma, 192 approved lncRNAs were extracted from HGNC (www.genenames.org). All the studies lncRNAs were then subjected into the cBioPortal dataset [9] and their alteration was analyzed in 179 samples of pancreatic adenocarcinoma (TCGA, provisional) whose RNA-seq v2 data was available. The lncRNAs which were altered in ≥10% of the patients were considered as candidate genes for further analysis. The expression level of these candidate lncRNAs were also examined in the studies tumorous samples. Using cBioPortal, the copy number variations including gene amplification or deletion were also examined in candidate lncRNAs to see if the altered expression arises from such genetic mutation. Using the lncR Inter dataset, the interaction of our candidate lncRNAs with DNA, RNA, transcription factors were evaluated [10]. The tendency of these candidate lncRNAs toward co-occurrence was examined using cBio Portal getaways. The contribution of candidate lncRNAs with pathologic parameters of the tumor samples like neoplasm disease stage, neoplasm histological grade, neoplasm histological type, primary tumor site and tumor anatomic site was also examined and analyzed in R using the cgdsr and cbaf [11]. The contribution of altered expression of candidate lncRNAs with patients’ overall survival was studied using Kaplan-Meier estimate. The Log-Rank Test P-Value <0.05 was considered as statistically significant. All the statistics were performed in R package.
Results
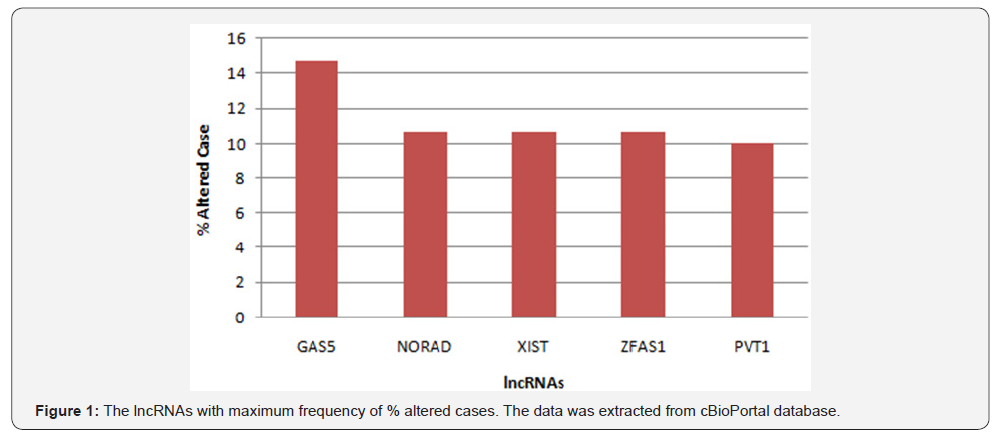
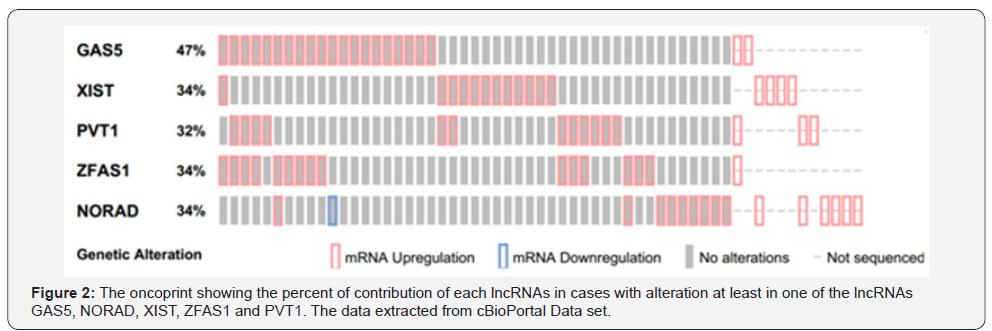
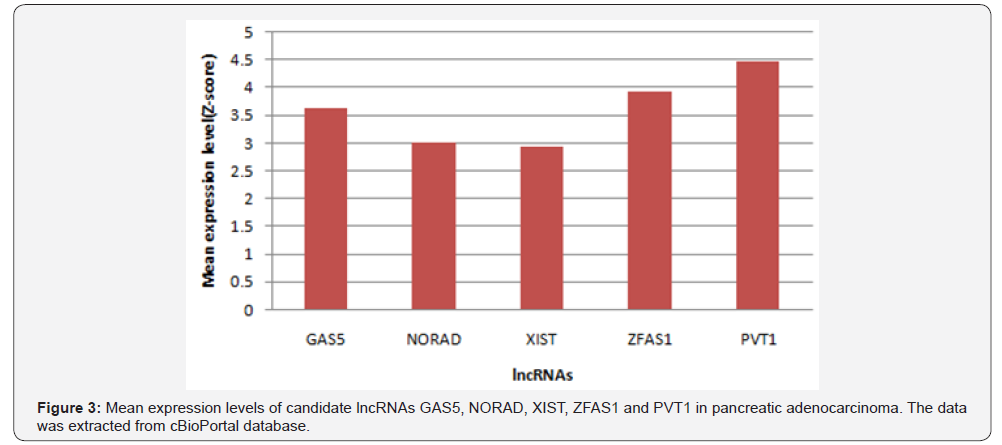
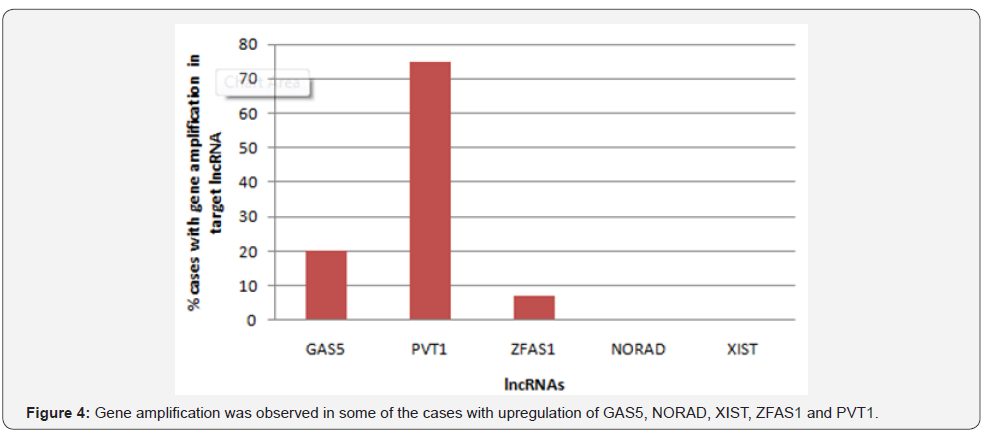
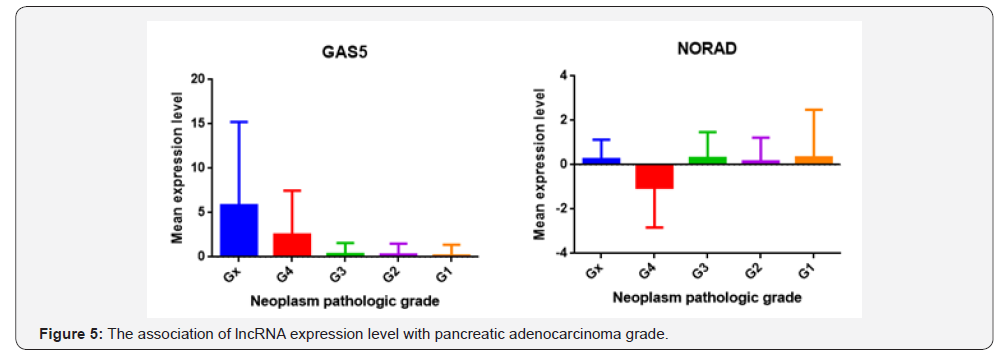
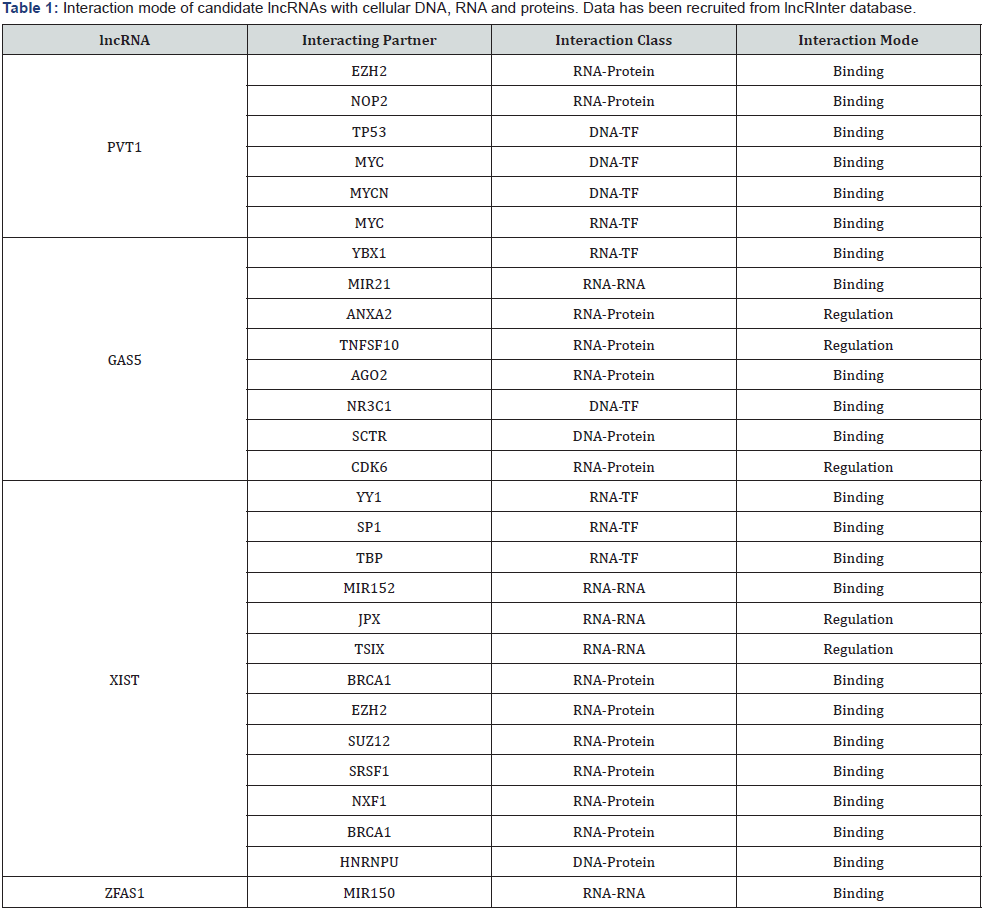
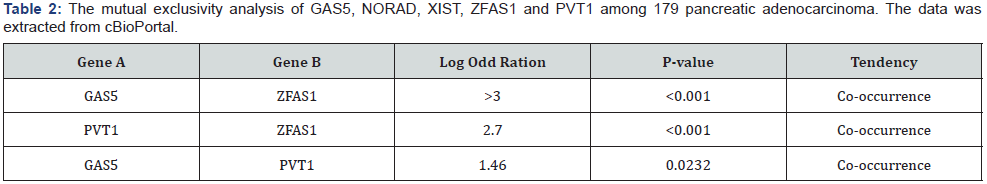
From 192 lncRNAs incorporated in this analysis, 59 lncRNAs have been altered at least in one patient. As shown in Figure 1, the lncRNAs GAS5, NORAD, XIST, ZFAS1 and PVT1 were altered in more than 10% of patients. Totally, 39% of the pancreatic adenocarcinoma cases altered for expression of at least one of these five lncRNAs and GAS5 was the gene which allocated the maximum number of cases with alteration in its expression level (Figure 2). We also found these five lncRNAs have been upregulated in cancer tissues with the Z-score more than three (Figure 3) in comparison with normal group. It was found that gene amplification was observed in 75%, 20% and 7% of cases who were upregulated for the lncRNAsPVT1, GAS5 and ZFAS1 respectively (Figure 4). As shown in Table 1, these lncRNAs interact with variety of gene sequence, transcripts and protein. We also found that GAS5, PVT1 and ZFAS1 have tendency toward co-occurrence (P-value<0.05) (Table 2). Inspite of NORAD, It was found that the expression level of all candidate lncRNAs was more upregulated in high grade tumor (G4) in comparison with low grade (G1) one although this value was statistically significant only in GAS5 (P-value=0.03) (Figure 5). In case of NORAD, we found that this lncRNA was downregulated in G4 grade tumor and increased expression was recorded in low grade samples (Figure 5).
Discussion
Here is the first report in which the contribution of lncRNAs with pancreatic adenocarcinoma was examined using available cancer bioportals. One hundred nighty two approved lncRNAs were considered in this study. The expression levels of these lncRNAs in pancreatic adenocarcinoma tissues as well as the percent of cases with alteration in lncRNAs were calculated separately. The lncRNAs with acceptable values in both analyses were considered as candidate lncRNAs. We found that lncRNA GAS5, NORAD, XIST, ZFAS1 and PVT1 were the genes with good score in both analyses and upregulated in tumorous tissues. In case of lncRNAs PVT1, GAS5 and ZFAS1, we found that this upregulation may partly due to gene amplification of the corresponding genes in some cases. We also found that GAS5, ZFAS1 and PVT1 have tendency toward co-occurrence which may due to presence of same regulator or contribution in same biological processes [3]. Our analysis showed that our lncRNAs interact with variety of cellular genes, RNAs and protein which some of these target partner including MYC, P53, miR-21, EZH2 play an important role in cell proliferation, growth, apoptosis and repair. Except NORAD lncRNA, all the other lncRNAs were upregulated in high grade samples showing the probable role of these genes in tumor invasion. However, NORAD was suppressed in high grade samples which propose this gene act in initiation step of pancreatic adenocarcinoma. The association of lncRNAs with pancreatic cancer has been recently reported in some studies [12]. Among which the role of lncRNAs MALAT1 [13], HOTAIR [14], MEG3 [15] and HOTTIP [16] in cell growth and progression of pancreatic cancer has been well- established. Similar to our obtained results, in a study by [17], they found that lncRNA PVT1 has been reported to be upregulated in pancreatic cancer cells and connected with poor prognosis in these patients. Zhao et al. [18] showed that PVT1 can promote the pancreatic cancer metastasis through an interaction with miR-448. The lncRNA NORAD has been recently reported in pancreatic cancer cell metastasis through the induction of EMT signaling pathways [19]. The oncogenic potential of XISTlncRNA has been demonstrated in pancreatic cancer which seem due to the interaction of this lncRNAs with miRNAs like miR- 133 and miR-34a [20,21]. However, mentioned study are limited to 1-2 studies and more experimental works is urgent to validate the involvement all of these lncRNAs in pancreatic cancer progression. In case of lncRNAs GAS5 and ZFAS1 no experimental works has been published until now. Regarding to above mentioned data, we strongly suggest that lncRNAs GAS5, NORAD, XIST, ZFAS1 and PVT1 are good choices for starting experimental project focus on lncRNAassociated pancreatic cancer which may open a new window I discovery of novel biomarker for this current cancer.
Acknowledgment
This study was affiliated to Ahvaz Jundishapur University of medical sciences. The authors are so thankful from all the colleagues whose comments improved the quality of this manuscript.
References
- Bhan A, Soleimani M, Mandal SS (2017) Long noncoding RNA and cancer: a new paradigm. Cancer Res 77(15): 3965-3981.
- Arshi A, Ansari H, Ghahramani Seno M, Doosti A, Khoramian M, et al. (2016) Comparison of GAS5 Long non-coding RNA Expression and NEAT1 in Breast Cancer Patients and Healthy People. Armaghane Danesh 21(3): 278-289.
- Birgani MT, Hajjari M, Shahrisa A, Khoshnevisan A, Shoja Z, et al. (2018) Long non-coding RNA SNHG6 as a potential biomarker for hepatocellular carcinoma. Pathol Oncol Res 24(2): 329-337.
- Ozawa T, Matsuyama T, Toiyama Y, Takahashi N, Ishikawa T, et al. (2017) CCAT1 and CCAT2 long noncoding RNAs, located within the 8q. 24.21 ‘gene desert’, serve as important prognostic biomarkers in colorectal cancer. Ann Oncol 28(8): 1882-1888.
- Shi T, Gao G, Cao Y (2016) Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis Markers 2016: 9085195
- Tempero MA, Arnoletti JP, Behrman S, Ben Josef E, Benson, et al. (2010) Pancreatic adenocarcinoma. J Natl Compr Canc Netw 8(9): 972-1017.
- Pandol SJ (2015) Normal pancreatic function. Pancreapedia: The Exocrine Pancreas Knowledge Base.
- Moschovis D, Gazouli M, Tzouvala M, Vezakis A, Karamanolis G (2017) Long non-coding RNA in pancreatic adenocarcinoma and pancreatic neuroendocrine tumors. Annals of gastroenterology 30(6): 622-628.
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2(5): 401-404.
- Liu CJ, Gao C, Ma Z, Cong R, Zhang Q, et al. (2017) lncRInter: A database of experimentally validated long non-coding RNA interaction. J Genet Genomics 44(5): 265-268.
- Birgani MT, Shahrisa A (2017) cbaf: Multiple automated functions for cbioportal.org. R package version 1.0.2.
- Huang X, Zhi X, Gao Y, Ta N, Jiang H, et al. (2016) LncRNAs in pancreatic cancer. Oncotarget 7(35): 57379-57390.
- Pang EJ, Yang R, Fu XB, Liu YF (2015) Over expression of long noncoding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol 36(4): 2403- 2407.
- Xien Z, Chen X, Li J, Guo Y, Li H, et al. (2016) Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget 7(18): 25408-25419.
- Hu D, Su C, Jiang M, Shen Y, Shi A, et al. (2016) Fenofibrate inhibited pancreatic cancer cells proliferation via activation of p53 mediated by upregulation of LncRNA MEG3. Biochem Biophys Res Commun 471(2): 290-295.
- Cheng Y, Jutooru I, Chadalapaka G, Corton JC, Safe S (2015) The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget 6(13): 10840-10852
- Huang C, Yu W, Wang Q, Cui H, Wang Y, et al. (2015) Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva Med 106(3): 143-149.
- Zhao L, Kong H, SunH, Chen Z, Che B, et al. (2017) LncRNA‐PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR‐448. J Cell Physiol 233(5): 4044-4055.
- Li H, Wang X, Wen C, Huo Z, Wang W, et al. (2017) Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxiainduced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol Cancer 16(1): 169.
- Wei W, Liu Y, Lu Y, Yang B, Tang L (2017) LncRNA XIST promotes pancreatic cancer proliferation through miR‐133a/EGFR. J Cell Biochem (10): 3349-3358.
- Sun Z, Zhang B, Cui T (2018) Long non-coding RNA XIST exerts oncogenic functions in pancreatic cancer via miR-34a-5p. Oncol Rep 39(4): 1591-1600.






























