Somatic Mutations in Cancer-Free Individuals: A Liquid Biopsy Connection
Andrew Ford, Charmaine Brown and Chen-Hsiung Yeh*
Circulogene Theranostics, USA
Submission: November 22, 2017; Published: January 09, 2018
*Corresponding author: Chen-Hsiung Yeh, Circulogene Theranostics, 3125 Independence Dr., Suite 301, Birmingham, AL 35209, USA, Tel: 1-205-234-0128; Email: cyeh@circulogene.com
How to cite this article: Andrew F, Charmaine B, Chen-Hsiung Y. Somatic Mutations in Cancer-Free Individuals: A Liquid Biopsy Connection. J Tumor Med Prev. 2018; 2(4): 555593. DOI: 10.19080/JTMP.2018.02.555593.
Abstract
Somatic mutations have been perceived as the causal event in the origin of the vast majority of cancers. Advanced massively parallel, high-throughput DNA sequencing have enabled the comprehensive characterization of somatic mutations in a large number of tumor samples for precision and personalized therapy. Understanding how these observed genetic alterations give rise to specific cancer phenotypes represents an ultimate goal of cancer genomics. However, somatic mutations are also commonly found in healthy individuals, which interfere with the effectiveness for cancer diagnostics.
Keywords: Somatic mutation; Germline; Cell-free DNA; Liquid biopsy; Next-generation sequencing
Mutations in Healthy Individuals are Not All Germline
Over the course of our lifetime, there are many millions of cell divisions in the body. By chance alone, mutations will definitely occur. Indeed, spontaneous somatic mutations constantly occur in individual cells. These background mutations arise either from replication errors or from DNA damage that is repaired incorrectly or left unrepaired, and have been detected in healthy tissues, including blood, skin, liver, colon, and small intestine[1-3]. Deep-sequencing studies in normal tissues also surprisingly identified cancer-driving mutations, e.g., in blood, driver mutations can be detected in ~10% of individuals older than 65 years of age and resemble patterns seen in leukemia patients. Individuals carrying these driver mutations have an elevated future risk of blood cancers [4-6], suggesting that these are genuine precancerous clones. Further, a detailed analysis of 31,717 cancer cases and 26,136 cancer-free controls from 13 genome-wide association studies revealed that the majority, if not all, of aberrations that were observed in the cancer-associated cohort were also seen in cancer-free subjects, albeit at lower frequency [7,8].
Over the course of our lifetime, there are many millions of cell divisions in the body. By chance alone, mutations will definitely occur. Indeed, spontaneous somatic mutations constantly occur in individual cells. These background mutations arise either from replication errors or from DNA damage that is repaired incorrectly or left unrepaired, and have been detected in healthy tissues, including blood, skin, liver, colon, and small intestine[1-3]. Deep-sequencing studies in normal tissues also surprisingly identified cancer-driving mutations, e.g., in blood, driver mutations can be detected in ~10% of individuals older than 65 years of age and resemble patterns seen in leukemia patients. Individuals carrying these driver mutations have an elevated future risk of blood cancers [4-6], suggesting that these are genuine precancerous clones. Further, a detailed analysis of 31,717 cancer cases and 26,136 cancer-free controls from 13 genome-wide association studies revealed that the majority, if not all, of aberrations that were observed in the cancer-associated cohort were also seen in cancer-free subjects, albeit at lower frequency [7,8].
Somatic Driver Mutations Found in Healthy Population by Liquid Biopsy
With the dramatically decreased cost of next-generation sequencing (NGS) in recent years, it is now practical to screen a large number of individuals at ultra-deep sequencing depths to identify cancer-related mutations. Cell-free DNA (cf DNA) in the blood circulation of cancer patients (as liquid biopsy) has emerged as key biomarkers for cancer monitoring and treatment decision-making [11]. Both academic research groups and industry players are chasing the pan-cancer screening by a simple blood draw. However, the reliable and accurate application of cfDNA detection requires better understanding of background somatic information in healthy individuals. as the variant allele frequency greater than 1% and the average depth more than 5,000 xs for demonstration. Our data revealed an age-independent mutation spectrum with average 3.12 somatic mutations per subject (Figure 1). The most frequently mutated genes are TP53 (42%), KIT (6%), KDR (5.5%), PIK3CA (5.5%), EGFR (5%) and PTEN (3.7%). These results highlighted the prevalence of some cancer-associated driver mutations in healthy individuals as background mutations.
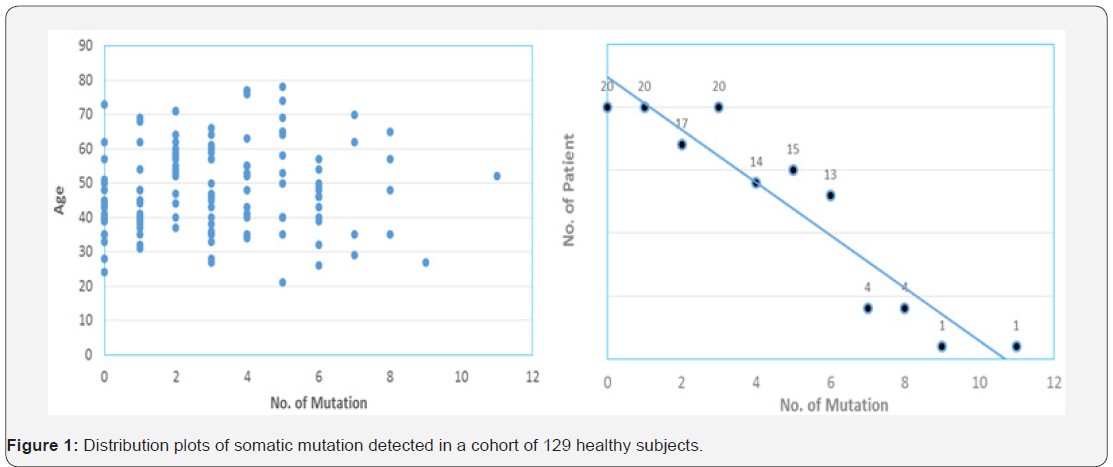
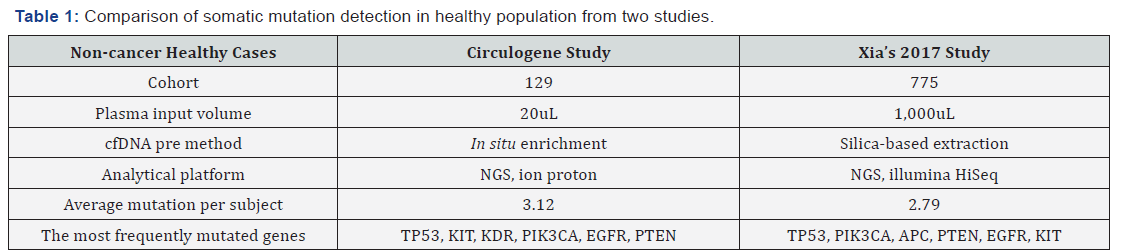
We also demonstrated the concordance between our results and a recent study for revealing the real somatic mutation in healthy population. The study by Xia et al. [12] examined the background somatic mutations in white blood cells and cfDNA in healthy controls based on sequencing data from 821 non-cancer individuals with the aim of understanding the baseline profile of somatic mutations detected in cfDNA. The data comparison was summarized in Table 1. Although there are differences in study cohort composition, sample volume, extraction methodology and analytical platform, the end results are remarkably similar, i.e., average 3 mutations per subject with an almost identical list of frequently mutated genes. Although varying mutation spectra in cancers have often been attributed to cancer-specific processes, our data suggest that at least a subset of these mutations actually reflect normal tissue-specific processes. This concept is consistent with the idea that a substantial fraction of the mutations found in cancers occur in normal stem cells [13,14].
Normal Tissue as a Germline Control not Justified
There is evidence for the presence of tumor-derived cfDNA in early cancers [15]. However, the real fraction of cfDNA that shed by tumor rather than the background somatic mutations is not well illustrated. For clinical application, the low level of tumor mutation as well as the heterogeneity of background mutation present in the circulation needs to be clearly addressed and differentiated to achieve accuracy. Unfortunately, this goal can’t be achieved by pushing detection limit of current advanced technology to below 0.01% mutant allele frequency (MAF). Contrarily, the higher sensitivity will guarantee higher chance to pick up background somatic mutations. Also, the clinical relevance of those low-percentage tumor mutations is still debatable in terms of treatment decision or regimen change.
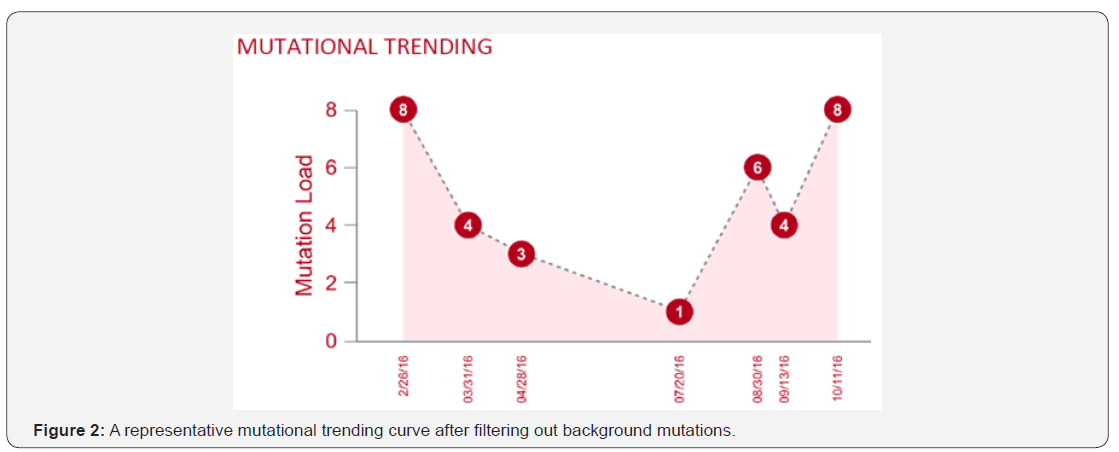
Each human individual is unique. Every cancer patient is different. No two tumors are the same even resides within the same patient; to distinguish the definitive cancer-specific mutations from background signals observable in plasma is extremely daunting. Evaluation of specificity in plasma cfDNA profiles from large numbers of healthy individuals as representative controls for the cancer population seems farfetched with uncertainty, especially when standardized protocol and optimized technology are still lacking. Unlike tissue genomic DNA, circulating cfDNA is so diluted and dynamic with a relatively short half-life, making single-point measurement not suitable for clinical application. We reason that cfDNA in circulation is truly under a continuous selection pressure to select for highly aggressive/proliferative clones, as disease progressing the low-abundant tumor clones will either evolve and dominate or vanish by the immune clean-up processes, therefore longitudinal clinical follow-ups should be performed to identify the best time and target for precision therapy, meanwhile to filter out contaminating background mutations. To achieve high clinical specificity, a cfDNA-based test must be capable of distinguishing between the background signals originating from non-cancer or pre-cancerous processes and the invasive malignancy of clinical interest. It is still possible that mutational signatures in cfDNA could distinguish basic biological processes from malignant and pathological processes. Here we propose a combined approach based on the tumor evolutional principle of “survival and domination of the fittest” in circulation that is to perform multiple time-point monitoring, filter out potential background mutations (e.g., <1% MAF), reduce sample input volume and interrogate multiple databases. A representative mutational trending curve following our approaches was shown (Figure 2).
Our findings underscore the importance of an assessment of the landscape of somatic mutations in cancer-free population, and associated mutation signatures. Somatic mutations and mosaicism in healthy individuals have implications not only for early detection, diagnosis and treatment of cancer using liquid biopsy but also emerging technologies in healthcare. We recommend caution while extending the mutation conclusions to cancer patients by employing matched normal tissue as germline control. To increase sample input and push liquid biopsy sensitivity toward <1% may not serve the interest of detecting low-frequency mutant allele, but only to increase the chance of background mutation contamination.
References
- Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, et al. (2016) Tissuespecific mutation accumulation in human adult stem cells during life. Nature 538(7624): 260-264.
- Martincorena I, Campbell PJ (2015) Somatic mutation in cancer and normal cells. Science 349(6255): 1483-1489.
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, et al. (2012) the origin and evolution of mutations in acute myeloid leukemia. Cell 150(2): 264-278.
- Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, et al. (2014) Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371(26): 2477-2487.
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, et al. (2014) Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371(26): 2488-2498.
- Mc Kerrell T, Park N, Moreno T, Grove CS, Ponstingl H, et al. (2015) Leukemia-associated somatic mutations drive distinct patterns of agerelated clonal hemopoiesis. Cell Rep 10(8): 1239-1245.
- Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, et al. (2012) Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet 44(6): 651-658.
- Forsberg LA, Absher D, Dumanski JP (2013) Non-heritable genetics of human disease: spotlight on post-zygotic genetic variation acquired during lifetime. J Med Genet 50(1): 1-10.
- Volik S, Alcaide M, Morin RD, Collins C (2016) Cell-free DNA (cfDNA): Clinical significance and utility in cancer shaped by emerging technologies. Mol Cancer Res 14(10): 898-908.
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, et al. (2015) Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348(6237): 880- 886.
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, et al. (1997) Presence of fetal DNA in maternal plasma and serum. Lancet 350(9076): 485-487.
- Xia L, Li Z, Zhou B, Tian G, Zeng L, et al. (2017) Statistical analysis of mutant allele frequency level of circulating cell-free DNA and blood cells in healthy individuals. Sci Rep 7(1): 7526.
- Tomasetti C, Vogelstein B, Parmigiani G (2013) Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc Natl Acad Sci USA 110(6): 1999-2004.
- Tomasetti C, Vogelstein B (2015) Cancer etiology: Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347(6217): 78-81.
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, et al. (2014) Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6(224): 224ra24.






























