Docking, Deformability, B-Factor, And Covariance Analysis Study of Cinnamaldehyde Dipeptidyl Peptidase-4 (Dpp4) Inhibitors To Generate the Anti-Diabetic Effect
Alhamyani Abdulrahman1*, Mohammad A Alrofaidi1, Alatawi Kahdr1, Asayel Alghamdi2, Yazan Alghamdi2, Abdulmajeed Alghamdi2, Mohammed Alghamdi2, Aljuri Alzahrani2 and Abdulaziz alzahrani1
1Pharmaceutical chemistry department, Faculty of clinical pharmacy, Al Baha University, Al Baha 65779 Saudi Arabia
2Faculty of clinical pharmacy, Al Baha University, Al Baha 65779 Saudi Arabia
Submission: December 20, 2023; Published: January 04, 2024
*Corresponding author: Alhamyani Abdulrahman, Pharmaceutical chemistry department, Faculty of clinical pharmacy, Al Baha University, Al Baha 65779 Saudi Arabia
How to cite this article: Alhamyani Abdulrahman*, Mohammad A Alrofaidi, Alatawi Kahdr, Asayel Alghamdi, Yazan Alghamdi, et al. Docking, Deformability, B-Factor, And Covariance Analysis Study of Cinnamaldehyde Dipeptidyl Peptidase-4 (Dpp4) Inhibitors To Generate the Anti-Diabetic Effect. JOJ Pub Health. 2024; 8(4): 555744. DOI: 10.19080/JOJPH.2024.08.555744
Abstract
Diabetes is among the most prevalent diseases around the globe and many drugs have been developed and tested based on different drug targets. However, natural products have become more attractive for drug discovery in the current situation. In the current study, we tested the potential of cinnamaldehyde compounds targeting dipeptidyl peptidase-4 (DPP4), a type II transmembrane glycoprotein that plays a vital role in the metabolism of glucose. DPP-4 inhibitors reduce glucagon and blood glucose levels. The mechanism of DPP-4 inhibitors is to increase incretin levels (GLP-1 and GIP), which inhibit glucagon release, which in turn increases insulin secretion, decreases gastric emptying, and decreases blood glucose levels. We conducted a molecular docking study and evaluated the stability of the complexes by molecular dynamics simulations and assessed their pharmacokinetic properties. The compound had a binding energy between -5.7 Kcal/mol. Moreover, the post molecular dynamics (MD) analysis indicated that they were all highly stable with DPP4 with comparable results. Pharmacokinetic parameters of the compounds showed brilliant properties. Our study suggested that compound could be active in treating DPP4 dysfunction to regulate glucose levels.
Keywords: Diabetes mellitus (DM); Dipeptidyl peptidase-4 (DPP4); Cinnamaldehyde
Introduction
Diabetes mellitus (DM) is increasing and is a leading worldwide health issue; especially type 2 DM [1]. The prevalence of diabetes mellitus (DM) is growing globally, and it has been estimated to touch 642 million marks by the end of 2040 (IDF-Diabetes Atlas 8thedn). It will be a serious health issue shortly because of an increase in the number of diabetes patients at an alarming rate [2]. Due to its various adverse effects and worsening clinical conditions, the existing therapy for the treatment of type 2 DM is not sufficient [3].
So, there is an urgent need to find novel breakthrough natural medicines due to the limitations of the currently available regimen for diabetes. The use of an integrated approach for the identification of novel inhibitors against a validated protein might be potentially desirable in this context. Among several proteins to target treatment for diabetes mellitus, dipeptidyl peptidase-4 (DPP4) protein is encoded by the DPP4 gene which is a membrane serine exopeptidase that functions by cleaving X-proline dipeptides from the N-terminus of polypeptides [4]. DPP4 is a serine protease that breaks down the peptidic hormone glucagon-like peptide 1 (GLP-1) which plays a significant role in regulating insulin release thereby controlling the level of blood glucose in the human body [5,6]. Various studies demonstrate that the inhibition of DPP4 could play a role in increasing incretin levels (GLP-1 and G IP), which inhibit glucagon release, which in turn increases insulin secretion, decreases gastric emptying, and decreases blood glucose [7,8]. Therefore, it has been considered a promising target for developing a novel drug for the treatment of type 2 diabetes.
Figure 1 The drug design and discovery are determined by developments in bioinformatics and several in silico tools that terminated in the transformation of ‘the bioinformatics era’ [9]. The in-silico method practices the grouping of ideas of computerized algorithms and statistical skills with advancements in high-performance computation in biological science [10]. So, bioinformatics and computational have ology have become the central pattern for fastmoving up the development and cost of searching new possible applicants for any targeted disease [11]. Binary key styles i.e., structure and ligand-based computergenerated showing methods have extensively been used to determine the compounds in spite of having some restrictions [12]. Combination of these in bioinformatics techniques appears to be a dependable method to identifying probable compound with improved therapeutic usages.
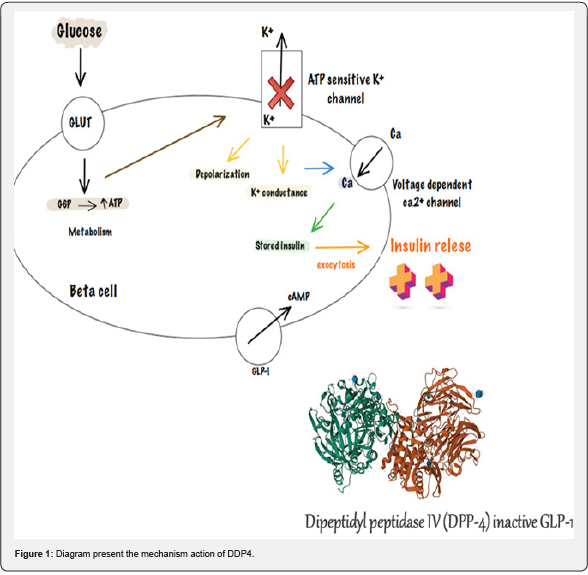
Since ancient times, some herbal remedies have been used to cure diabetes, manage blood sugar levels, and prevent insulin resistance, which is the leading cause of type 2 diabetes, [13]. In India, Cinnamomum zeylanicum (cinnamon) is widely used in traditional medicine to treat diabetes [14]. Cinnamaldehyde, one of the active ingredients derived from cinnamon, has been used in the kitchen and industry as a natural flavoring and fragrance agent. Cinnamaldehyde, an old flavorant derived from cinnamon trees and other species of the genus Cinnamomum, is now gaining popularity due to its ability to prevent the development of diabetes and its complications [15]. were selected for identifying their potential efficacy against DPP4 protein in this study.
Materials and Methods
Preparation of compound and receptor
The compound named cinnamaldehyde was carefully chosen for this search. The 3D structures of the compound selected were drawn in ChemDraw Ultra 12.0 [16] and their canonical SMILES were generated. Then they were transformed into protein data bank files (PDB) via Online SMILES Translator and Structure File Generator (https://cactus.nci.nih.gov/translate/). Crystal assembly of human dipeptidyl peptidase IV (DPP4) in complex with a decapeptide (tNPY) at 2.3 Å resolution was available in Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) [17]. The structure was composed and completely the ligand molecules, hetero atoms, and water molecules were eliminated from the structure utilizing the program PyMOL to visualize and analyze protein structures [18].
Molecular docking simulation of cinnamaldehyde compound with DPP4
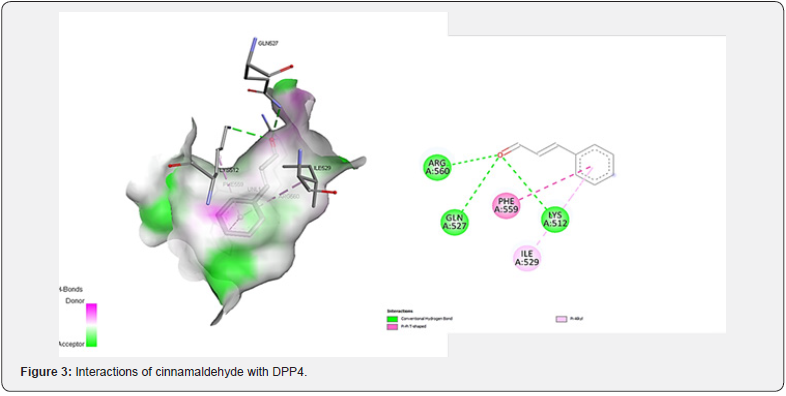
Molecular docking was applied via PyRX [19], an open-source simulated screening software program to check receptor-ligand interactions. The significant catalytic site residues A:560 (ARG), A:527 (GLN), A:512 (LYS), A:559 (PHE), and A:529 (ILE) were surrounded with the parameters of the grid box with X=23, Y=23, Z=23 (Center grid box: X = 60.331, Y = 10.448, Z = 21.064) dimensions. AutoDock Vina (ADV) [20], implemented in PyRX was utilized to achieve all the docking simulations with the predetermined parameters. Extra, the protein-ligand interaction was abstract by Pymol 1.1 [21].
Molecular Dynamic Simulation Deformability, B-factor, and Covariance
We use Desmond software to perform 100 nanoseconds of molecular dynamics simulation [22]. The input complex for the simulation was taken from docking results. Docking gave binding analysis of protein and ligand in rigid and static mode [23]. While molecular dynamics simulation predicts the protein and ligand interaction and energies through the time of simulation in physiological environment [24,25]. Protein Preparation Wizard of Maestro was used to preprocess, optimize, and minimize the protein-ligand complex. After that System Builder was used to prepare complex for simulation. We used TIP3P (Intermolecular Interaction Potential 3 Points Transferable) solvent model, the shape of grid was orthorhombic the temperature was 310 K with 1 atm pressure and OPLS_2005 force field was used. We add 0.15 M sodium chloride to mimic the physiological environment and add ions to neutralize the system. While adding ions we create an excluded region of 20 Angstrom for ligand, so that the ion should not disturb the interactions of protein with ligand in simulation. We stored the trajectories after every 100 ps in 100 ns of simulation [26]. Principal component analysis (PCA) and dynamic cross-correlation matrix (DCCM) were analyzed by using ‘Bio3D’ package of R [27].
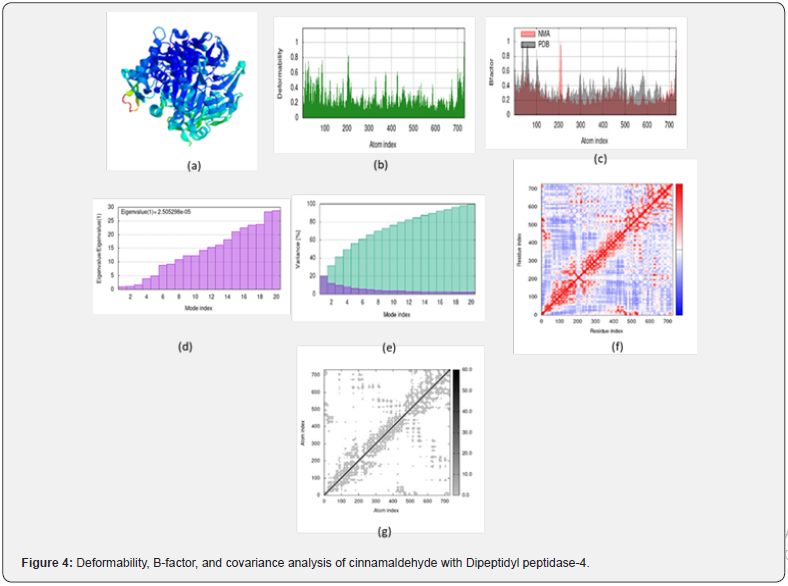
Dynamics simulation of molecule evaluation was conducted on the best-designated cinnamaldehyde -DPP4 study performed via iMODS server (http://imods.chaconlab.org/). iMODS can be utilized professionally to examine the structural dynamics of the protein complexes so that will be fast and easy to server. The software program offers the standards of deformability, B-factor (movement outlines), eigenvalues, variance, co-variance map, and elastic network, for a protein complex. The deformability is contingent on the capability to warp at each of its amino acids. The eigenvalue is associated with the dynamics that is needs to deform structure and the lower eigenvalue matches to the easier deformability of the complex. Eigenvalue can represent the wave stiffness of the protein complex. In addition to defining and calculating protein flexibility [28-30]. For analyzing the molecular dynamics simulation of the cinnamaldehyde with DPP4 docked complex was used.PDB files were uploaded to the iMODS software program, and the results were shown keeping all the parameters as defaulting.
ADME analysis
SwissADME [31] was utilized to evaluate the pharmacokinetics properties of the cinnamaldehyde compound.
Results
Molecular Docking Analysis of Cinnamaldehyde with Dipeptidyl peptidase-4
AutoDock 4.2 detected five probable binding sites for the cinnamaldehyde compound as output. The good position selected for the compound depends on the lowest docking energy. The docking energy score of the cinnamaldehyde compound Figure 2 with DPP4 was 5.7 Kcal/mol. The amino acid interactions of DPP4 with the compound were also recognized. The cinnamaldehyde compound established 3 hydrogen bonding interactions with the protein LYS512, GLN527 and ARG560 were common interacting residues Figure 3.
Molecular Dynamics Simulation of Cinnamaldehyde with Dipeptidyl peptidase-4
Figure 4 shows the results of molecular dynamics simulation and the evaluation of the normal mode analysis (NMA) to cinnamaldehyde plus the Dipeptidyl peptidase-4 docked complex. Deformability graph designates the high level of peaks which match to the areas of the protein with potential deformability Figure 4b. B-factor graph of the complex indicates informal imagining of the alteration also a contrast between the NMA and the PDB field of the docked complex Figure 4c. The eigenvalue of the complex is demonstrated in Figure 4d. cinnamaldehyde and Dipeptidyl peptidase-4 docked complex generated an eigenvalue of 2.505298e-05. The variance graph shows the separate alteration as remarkable in red colored column and cumulative variance via green colored column Figure 4e illustrate co-variance map of the complex where the associated wave between a pair of residues is designated through red color, unassociated wave is showed via white color and anti-associated wave is specified thru blue color.
The elastic map of the complex characterizes the joining between the atoms and darker gray zones match to the stiffer zones Figure 4g [28- 30]. Dynamic activities of atoms and conformational differences of Cα atoms of the protein-ligand complexes were measured by RMSD to distinguish their stability. It is shown that all the complexes display lesser RMSD (<0.4 nm) without common fluctuations representative their superior stability revealed in Figure 5. After some minor fluctuations until 60ns, all the complexes retained stability till the end of the simulation period. Flexibility of separate residue was considered in relations of RMSF to get a well insight into the region of proteins that are presence fluctuated during the simulation. Understood the binding of compounds makes the protein a little flexible in 200-300 and 650-700 residue areas Figure 5.

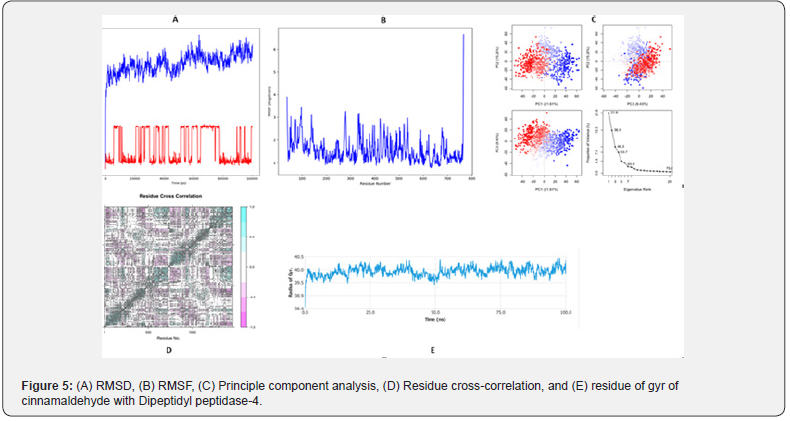
The density of the complex was characterized by the radius of gyration (Rg). A lower degree of fluctuation during the simulation period shows the better density of a system. The Rg of the complexes was found to be lower than in the beginning period while the DPP4- cinnamaldehyde complex was the most compact Figure 5. Interaction between protein-ligand complexes and solvents was calculated by solvent-accessible surface area (SASA) over the simulation period. So, the SASA of the complex was measured to analyze the extent of the conformational variations that happened during the interaction. Remarkably, the protein highlighted an insignificant decrease of the surface area showing a moderately minor SASA calculation than the initial period Figure 5. Hydrogen bonding between a protein-ligand complex is crucial to steady the structure. It was shown that the maximum number of conformations of the protein formed up to two hydrogen bonds with the DPP4 Figure 5.
ADME analysis
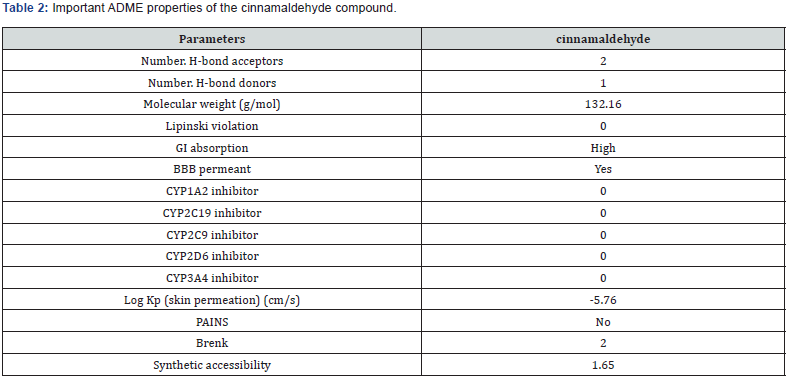
The ADME advantages of the cinnamaldehyde compound were projected using the Swiss ADME server Table 1. It has been showing significant value. Interestingly, do violate Lipinski’s rule of five and they have high GI absorption. The cinnamaldehyde compound does not reduce any isoform of the cytochrome P450 enzyme. The log kp (skin diffusion) was in good range and they also had efficacy in crossing the blood-brain barrier. The PAINS alert confirmed the absence of catechol moieties in the compounds and the synthetic accessibility value was also low. The cinnamaldehyde compound showed good outcomes in both binding with the protein and drug-likeliness properties. Considering binding affinities, interactions with target proteins, and ADMET profile data, we have analyzed protein-ligand interaction complexes using a molecular docking approach and found exciting results. These results are valuable references for supplementary investigational research in designing new forceful DPP4 inhibitors Table 2.

Discussion
Computer-aided drug design (CADD) methods are currently utilized professionally in drug discovery and development courses besides traditional methods. Among several approaches of CADD that are evaluated as favorable techniques, molecular docking, and molecular dynamics simulations are mostly utilized to perform virtual screening of compounds to analyze how the ligands inhibit a specific target protein. Also, the pharmacological properties of the cinnamaldehyde compound can be assessed using the increasing development of robust computational tools before entering into the experimental trial. So, intensive CADD approaches were performed in this study to assess the drug-gable properties of the cinnamaldehyde compound against the DPP4 protein.
Our study, we focused on both the binding affinity and ADMET characterizations of the cinnamaldehyde compound to test their efficacy against DPP4 which is an intrinsic membrane glycoprotein playing a major role in glucose metabolism and provides a useful treatment for diabetes mellitus type 2. For this, the cinnamaldehyde compound was selected and docked against the DPP4. It was shown that cinnamaldehyde displays the highest binding affinity of -5.7 kcal/mol while binding with the DPP4 and interacted with the highest number of amino acid residues in the active site. Furthermore, the interaction analysis highlights the potential of residues B:560 (ARG), B:527 (GLN), B:510 (PRO), B:559 (PHE), B:529 (ILE) and B:512 (LYS) in the binding of the DPP4.
The ADME properties of the cinnamaldehyde compound describe high gastrointestinal absorption potency with a skin permeation coefficient (Log Kp) of 5.76. No compounds violate Lipinski’s rule of five, thus, they might easily mimic the native ligand. Also, results of blood-brain barrier membrane permeability reveal that the cinnamaldehyde compound can cross the bloodbrain barrier. The Pan-assay interference compound (PAINS) is the cinnamaldehyde compound that tends to give incorrect optimistic results in high-throughput screens reacting nonspecifically with various biological targets. No compounds possessed any catechol group revealed from the PAINS filter. It has a low score of synthetic accessibility denoting that these compounds can be synthesized much more easily in experimental laboratories.
In this study has been conducted through in-silico analysis, it could be some limitations. The cinnamaldehyde compound, although they showed tremendous results with DPP4 protein, is yet to be performed in an animal model experiment and MD. So, acceptable results validations are needed more experimental to ensure their therapeutic effectiveness.
Conclusion
Potentiality of cinnamaldehyde compound targeting dipeptidyl peptidase-4 (DPP4). DPP-4 inhibitors decrease glucagon and blood glucose levels by rise incretin levels (GLP- 1 and GIP), which inhibit glucagon release, as result insulin secretion, decreases gastric emptying, and decreases blood glucose levels. We performed molecular docking analysis, tested the complexes in static molecular docking, and explored their pharmacokinetic properties. Our research indicates that cinnamaldehyde compounds, with a binding energy of -5.7 Kcal/ mol, have the potential to treat DPP4 dysfunction and regulate glucose levels. However, further testing on animal experiments is required to confirm their therapeutic effectiveness..
Acknowledgement
I would like to thank all colleagues who contributed to this study.
Conflict of Interest
None
References
- Aguiar C, Duarte R, Carvalho D (2019) New approach to diabetes care: From blood glucose to cardiovascular disease. Rev Port Cardiol English Ed 38: 53-63.
- Shaw JE, Sicree RA, Zimmet PZ (2010) Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res Clin Pract 87(1): 4-14.
- Gupta R, Walunj S, Tokala R, Kishore VLP, Santosh KS, et al. (2009) Emerging drug candidates of dipeptidyl peptidase IV (DPP IV) inhibitor class for the treatment of Type 2 Diabetes. Curr Drug Targets 10: 71-87.
- Darmoul D, Rouyer FC, Blais A, Voisin T, Sapin C, et al. (1991) Dipeptidyl peptidase IV expression in rat jejunal crypt-villus axis is controlled at the mRNA level. Am J Physiol 261(5 Pt 1): G763-9.
- Sebokova E, Christ A, Boehringer M, Jacques M, et al. (2007) Dipeptidyl Peptidase IV Inhibitors: The Next Generation of New Promising Therapies for the Management of Type 2 Diabetes. Curr Top Med Chem 7: 547-555.
- Galstyan KO, Nedosugova L V, Petunina NA, Julia AT, Natalia VV, et al. (2015) First Russian DPP-4 inhibitor Gosogliptin comparing to Vildagliptin in type 2 diabetes mellitus patients. Diabetes Mellit 19: 89-96.
- Zander M, Madsbad S, Madsen JL, Jens JH (2002) Effect of 6-week course of glucagon-like peptide 1 on glycaemic control, insulin sensitivity, and beta-cell function in type 2 diabetes: a parallel-group study. Lancet 359: 824–830.
- Prajapat R, Bhattacharya I (2016) In-silico Structure Modeling and Docking Studies Using Dipeptidyl Peptidase 4 (DPP4) Inhibitors against Diabetes Type-2. Adv Diabetes Metab 4: 73-84.
- Bruno A, Costantino G, Sartori L, Marco R, (2019) The in Silico Drug Discovery Toolbox: Applications in Lead Discovery and Optimization. Curr Med Chem 26: 3838-3873.
- Ekins S, Mestres J, Testa B (2007) In silico pharmacology for drug discovery: methods for virtual ligand screening and profiling. Br J Pharmacol 152(1): 9-20.
- Shaker B, Ahmad S, Lee J, Chanjin J, Dokyun N, et al. (2021) In silico methods and tools for drug discovery. Comput Biol Med 137: 104851.
- Aparoy P, Reddy KK, Reddanna P (2012) Structure and Ligand Based Drug Design Strategies in the Development of Novel 5-LOX Inhibitors. Curr Med Chem 19: 3763.
- Jeremiah O, Sogolo L (2020) Antioxidant effects and mechanisms of medicinal plants and their bioactive compounds for the prevention and treatment of Type 2 Diabetes: An Updated Review. Oxid Med Cell Longev 2020: 1356893.
- Subash Babu P, Prabuseenivasan S, Ignacimuthu S (2007) Cinnamaldehyde--a potential antidiabetic agent. Phytomedicine 14(1): 15–22.
- Zhu R, Liu H, Liu C, Wang L, Ma R, et al. (2017). Cinnamaldehyde in diabetes: A review of pharmacology, pharmacokinetics and safety. Pharmacological research 122: 78–89.
- Cousins KR (2011) Computer review of ChemDraw Ultra 12.0. J Am Chem Soc 133(21): 8388.
- Zardecki C, Dutta S, Goodsell DS, Maria V, Stephen K, et al. (2016) RCSB Protein Data Bank: A Resource for Chemical, Biochemical, and Structural Explorations of Large and Small Biomolecules. J Chem Educ 93: 569–575.
- Schrödinger L, DeLano W PyMOL (2020).
- Dallakyan S, Olson AJ (2015) Small-molecule library screening by docking with PyRx. Methods Mol Biol 1263: 243-250.
- Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31: 455.
- Rigsby RE, Parker AB (2016) Using the PyMOL application to reinforce visual understanding of protein structure. Biochem Mol Biol Educ 44: 433-437.
- Bowers KJ, David E, Xu Huafeng, Dror Ron O, and Eastwood MP, et al. (2006) Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. SC '06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. IEEE 43.
- Ferreira LG, Ricardo NDS, Glaucius O, Adriano DA, et al. (2015) Molecular docking and structure-based drug design strategies. Molecules 20(7): 13384-13421.
- Hildebrand PW, AS Rose, JKS Tiemann (2019) Bringing Molecular Dynamics Simulation Data into View. Trends Biochem Sci 44(11): 902-913.
- Rasheed MA, Muhammad A, Iqra A, Falak SK, Sumaira K, et al. (2021) Identification of Lead Compounds against Scm (fms10) in Enterococcus faecium Using Computer Aided Drug Designing. Life (Basel) 11(2): 77.
- Shivakumar D, Joshua W, Yujie W, Wolfgang D, John S, et al. (2010) Prediction of Absolute Solvation Free Energies using Molecular Dynamics Free Energy Perturbation and the OPLS Force Field. Journal of Chemical Theory and Computation 6(5): 1509-1519.
- Grant BJ, L Skjaerven, XQ Yao (2021) The Bio3D packages for structural bioinformatics. Protein Sci 30(1): 20-30.
- Prabhakar PK, Srivastava A, Rao KK, Balaji PV (2016) Monomerization alters the dynamics of the lid region in Campylobacter jejuni CstII: an MD simulation study. J Biomol Struct Dyn 34: 778-791.
- López Blanco JR, Aliaga JI, Quintana Ortí ES, Chacón P (2014) iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res 42: W271–276.
- Lopéz Blanco JR, Garzón JI, Chacón P (2011) iMod: multipurpose normal mode analysis in internal coordinates. Bioinformatics 27: 2843-2850.
- Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Reports 7: 42717.






























