Isothermal Amplification Assay for Detection of Chilli Leaf Curl Virus Infecting Chilli and its Vector Whitefly Cryptic Species
Mahmoud Omar Hassan1* and Sanad M Alsobeai2
1ICAR-Indian Institute of Horticultural Research, Hessaraghatta Lake PO, Bangalore-560089, Karnataka, India
2Department of Plant Pathology, Sri Venkateswara Agriculture College, Tirupati-517502, Acharya N G Ranga Agricultural University, Andhra Pradesh, India
Submission:September 19, 2024;Published: October 08, 2024
*Corresponding author: Venkataravanappa, Department of Plant Pathology, Sri Venkateswara Agriculture College, Tirupati-517502, Acharya N G Ranga Agricultural University, Andhra Pradesh, India
How to cite this article: Navyatejaswini B, Venkataravanappa V, Sarada Jayalakshmi R, Madhavi Reddy K, Chowdappa A, et al. Isothermal Amplification Assay for Detection of Chilli Leaf Curl Virus Infecting Chilli and its Vector Whitefly Cryptic Species. JOJ Hortic Arboric. 2024; 4(5): 555650. DOI: 10.19080/JOJHA.2024.04.555650.
Abstract
Leaf curl disease of Chilli is a major limiting factor for cultivation of chilli crop in different location of world including Indian subcontinent. The disease is known to cause by whiteflies transmitted begomoviruses complex leads to yield loss upto 100 percent in severe cases. PCR is the most commonly used diagnostic method for detection of begomovirus in both chilli and its whitefly cryptic species. However, the technique needs more time, high end equipments and laboratory conditions and major limitation is lack of field applicability. Therefore, loop-mediated isothermal amplification assay (LAMP) is more sensitive then PCR and have field applicability in quicker detection of chilli leaf curl virus (ChLCV) in chilli and its whitefly cryptic species. LAMP was carried out by targeting the Rep gene (AC1) of the virus which resulted in a laddered like amplification pattern were visualized in agarose gel electrophoresis. Further the above assay was validated by testing ChLCV infected chilli samples and other begomovirus infected samples revealed that only positive amplification was observed in ChLCV infected samples not for other crops begomviruses infected samples. This assay was further extended to detect the virus in whiteflies collected from the chilli fields, which is very essential to assess presence of virus in vector before the crop planting in the field. Thus, LAMP assay developed in this study is very useful in rapid diagnosis of ChLCV infections in chilli and its whitefly cryptic species.
Keywords:Begomovirus; Leaf curl, PCR, LAMP assay; Whitefly cryptic species; Tobacco Leaf Curl Virus (TbLCV) from tobacco; Okra Leaf Curl Virus (OLCV) from okra and tomato leaf curl virus (ToLCV) from tomato
Abbreviations:ChLCV: Chilli Leaf Curl Virus; TbLCV: Tobacco Leaf Curl Virus; OLCV: Okra Leaf Curl Virus; RCA: Rolling Circle Amplification; OW: Old World; ORFs: Open Reading Frames
Introduction
Chilli (Capsicum annum L.) is an important spice cum vegetable crop of the family Solanaceae [1] cultivated in different parts of India. The production crop is threatened by different biotic and abiotic stress from germination to harvest. Of these viruses are most economically important pathogens in causing yield loss up to 100% at different stage of infection during the crop growth period. More than sixty five different viruses have been reported infecting chilli crops in different parts of the world [2]. Chilli leaf curl disease, predominantly caused by different types of begomoviruses are most prevalent pathogen in India concerning both incidence and yield loss, particularly in severe cases [3]. Chilli leaf curl virus (ChLCV) is transmitted by whitefly cryptic species in various parts of the world in a semi-persistent manner, the infected plants showed a diverse range of symptoms under field condition [4,5].
Universal primers are commonly employed for PCR-based diagnosis of DNA viruses due to their high sensitivity and specificity. However, this technique necessitates expensive equipment, rendering it impractical in field settings where portable instruments are lacking. Furthermore, the lengthy PCR detection process, spanning several hours, adds to its limitations. This method’s infeasibility for field use is exacerbated by the high cost of equipment and the absence of suitable portable instruments. Consequently, there is a pressing need for a swift and straightforward assay for routine virus diagnosis in the infected plants. To address these challenges, isothermal amplification methods have emerged as an alternative, aiming to reduce costs and enhance the accessibility of PCR-based testing. These methods exhibit improved sensitivity and specificity compared to ELISA-based approaches [6]. Notably, LAMP assay has gaining its popularity for detecting plant pathogens across various crops [7].
LAMP is a novel gene amplification technique, offers rapidity, simplicity, and high specificity [8]. The LAMP reaction requires six specially designed primers and DNA polymerase. The advantage of LAMP is its ability to directly amplify specific DNA sequence under isothermal conditions, and most importantly, this method does not require a denatured DNA template [9]. The LAMP-based assay has been extensively utilized for detecting DNA viruses in various crops [6,7,10-13]. This study discusses the efficient LAMP-based assay for detection of chilli leaf curl virus (ChLCV) in chilli and its vector whitefly cryptic species.
Materials and Methods
Collection of virus infected chilli samples and whitefly cryptic species
During year 2022, survey was conducted for the collection of leaf samples of chilli showing typical leaf curl symptoms from Guntur (District), Andhra Pradesh which is vast chilli growing area of the state. Total ten samples showing diverse kind of symptoms was collected from different farmer’s field along with one non-symptomatic sample in same location. Similarly, we gathered ten samples of whiteflies in each location, comprising a minimum of 10 to 50 adult whiteflies directly from the fields using an aspirator. Then collected whiteflies were placed in 1.5- mL Eppendorf cylindrical tubes was labelled and transported to the Entomology laboratory. Subsequently, the whitefly specimens underwent for morphological identification by examining the fourth instars nymphs under a microscope [14]. Then the collected whiteflies were anesthetized and placed in a 2-mL Eppendorf tube containing 70% ethanol. Then tubes were stored at 4°C for further use.
DNA isolation, PCR detection and Sequencing
The total DNA isolated from ten chilli samples showing diverse kind of symptoms along with healthy chilli leaf sample using CTAB [15]. The DNA concentration was measured using Nanodrop separately (ND-1000 Spectrophotometer- V3.2) according to the manufacturer’s protocols. The purity of isolated DNA was checked by obtaining the mean absorbance value (A260/A280) of 1.5 to 1.7. Further the quality of the DNA was analyzed by electrophoresis through 0.8% agarose gel using 0.5 x TBE as running buffer. DNA bands were visualized with a UV transilluminator after gels were stained with ethidium bromide. The 50ng of the pure DNA of the ten chilli samples was subjected to PCR amplification to detect the begomovirus infection in chilli samples using begomovirus-specific primers pair designed to amplify genome segment of DNA A (Forward primer 5’-GCTCCCTGAATGTTCGGATGGA-3’ and Reverse primer -GTTCTCRTCCATCCATATCTTAC-3’) [16]. Further complete genome of virus was amplified by rolling circle amplification method (RCA) as described by Venkataravanappa et al. [17]. The amplified RCA (2 μl) product was digested with BamH1 for DNA-A and the linearized RCA fragment of 2.7 kb was cloned into pUC19 cloning vector (Thermo Fisher Scientific, Carlsbad, CA) and confirmed clones were sequenced by primer walking method by using universal M13 primers at Meaduxin Genomics India Pvt. Ltd, Bangalore, India.
Whitefly DNA Isolation
The total genomic DNA of whiteflies was extracted using HiGeno MB- HiPura insect DNA extraction kit as per the manufacturer’s protocol (Himedia, India). The DNA concentration was measured as described above. Further the presences of begomovirus in whiteflies were detected by PCR using specific primers (Forward primer 5’-GCTCCCTGAATGTTCGGATGGA-3’ and Reverse primer -GTTCTCRTCCATCCATATCTTAC-3’) [16].
LAMP assay
LAMP primer designing
Rep protein (AC1) gene sequence of ChLCV was used to design the specific LAMP primers using the Primer Explorer V.5 software (https://primerexplorer.jp/e) program. The specificity of the LAMP primers was confirmed through BLASTn search (Standard nt BLAST available at https://blast .ncbi.nlm.nih/gov/blast.cgi) (Table 1, Supplementary Figure 1).

Detection of ChLCV by PCR using LAMP primers in chilli and whitefly cryptic species
The PCR amplification was performed by using outer LAMP primers F3 and B3. The PCR amplification was executed using the Gene Amp PCR system 9700 (PE Applied Biosystems in Foster City, CA) in volumes of 25 μl PCR mixture as described by Venkataravanappa et al. [18]. LAMP amplified products was stained with ethidium bromide (10 mg/ml) and visualized by using Gel documentation system (Alpha Innotech, USA).
LAMP assay
The LAMP assay involved the utilization of template DNA extracted from infected chilli sample and the whitefly vector. A 20 μl LAMP-PCR reaction mixture was prepared, comprising 10 μM each of ChLCV-F3, ChLCV-B3, ChLCV-FIP, ChLCV-BIP, ChLCV-LF, and ChLCV-LB primers. The LAMP assay was performed as described by Venkataravanappa et al. [12]. Subsequently, the amplified LAMP products were analyzed on a 2% agarose gel, subjected to 65 V for 2 hours, and visualized using ethidium bromide stain, observed under a Gel documentation system (Alpha Innotech, USA).
Visual detection
The amplified LAMP products were visually with the help of 20 mM of hydroxy naphthol blue (HNB) colouring agent (Lemongreen, Shanghai, China) added separately to the LAMP master mix before to the PCR amplification. A change in colour from violet to sky blue indicates positive reactions.
Validation of LAMP assay
To validate the synthesised LAMP primers, total DNA isolated from ChLCD infected samples and whiteflies collected from Warangal (Telangana), Raichur (Karnataka), Jalna (Maharashtra) and Indore (Madhya Pradesh) state of India along with DNA extracted from the positive cultures of tobacco leaf curl virus (TbLCV) from tobacco, okra leaf curl virus (OLCV) from okra and tomato leaf curl virus (ToLCV) from tomato were included LAMP assay.
Results
PCR assay for detection of begomovirus in Chilli and whitefly cryptic species
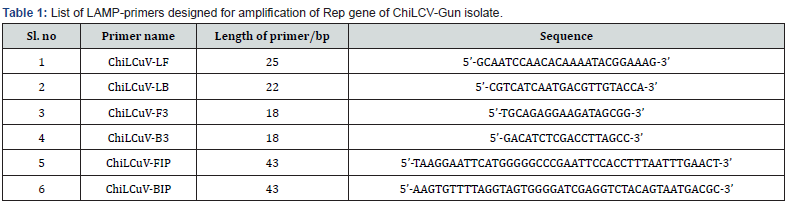
The chili samples exhibiting leaf curl symptoms collected from Farmers fields at Guntur and its vector whitefly cryptic species were confirmed to have begomovirus infection through partial genome amplification (1.2 kb) (Figure 2a) and sequenced in both chili sample and whitefly vector. Sequence analysis showed that all ten chilli samples showed more homology (more 96 % nucleotide identity) with already reported chilli leaf curl virus in both chilli and its vector whitefly cryptic species, indicating they are Belongs to single species. Therefore, one representative a sample was used to amplify the complete viral genome using the RCA method, cloned, and sequenced.
The viral genome sequence (ChLCV-Guntur isolate) analysis was done by utilized different omics programs (Clustal X2, Sea View, and Bioedit). The resulting consensus sequence was submitted to the Genbank database under the following accession number OR666666. The viral genome, spanning 2609 nucleotides, exhibits a genome structure akin to that of a monopartite begomovirus from the Old World (OW) and its encodes five conserved open reading frames (ORFs): two ORFs (V2 and V1) on the sense strand and four ORFs (C1, C2, C3, C4) on the antisense strand of the DNA component. The DNA-A component of ChLCV-Guntur isolate was compared with selected begomoviruses available in the GenBank. The result revealed the ChLCV-Guntur isolate is showed maximum nucleotide identity of 99% with ChLCV-infecting chilli (MN417112) in Andhra Pradesh, in which the sequence is available in the GenBank.
LAMP assay and Visual detection
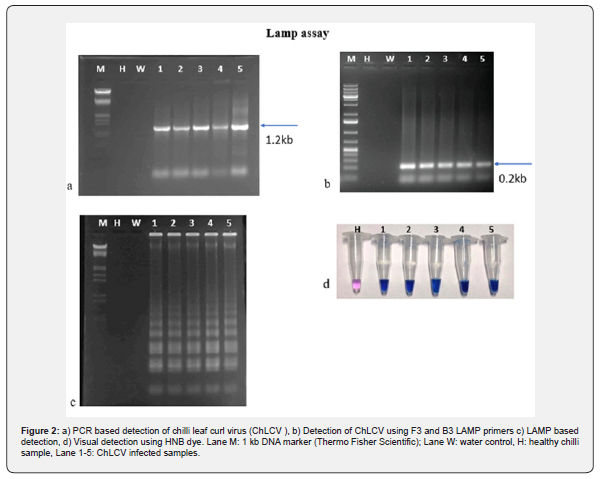
To detect ChLCV in chili samples collected from Guntur district of Andhra Pradesh using the LAMP assay, the samples were first subjected to PCR amplification with LAMP primers (ChLCV-F3 and ChLCV-B3). All reactions showed positive results with a 180 bp of PCR amplicon observed on an agarose gel (Figure 2b). Subsequently, six sets of LAMP primers were used to carry out the assay. The results demonstrated that all the infected samples displayed positive reactions with a ladder-like pattern (Figure 2c), while no amplification was observed in the healthy and water control samples on the 2% agarose gel. The positive reactions were easily visualized with the naked eye, as the positive samples exhibited a sky-blue color pattern, whereas the negative and water control samples showed a violet color when treated with Hydroxy naphthol blue dye before PCR amplification (Figure 2d).
Sensitivity of LAMP assay
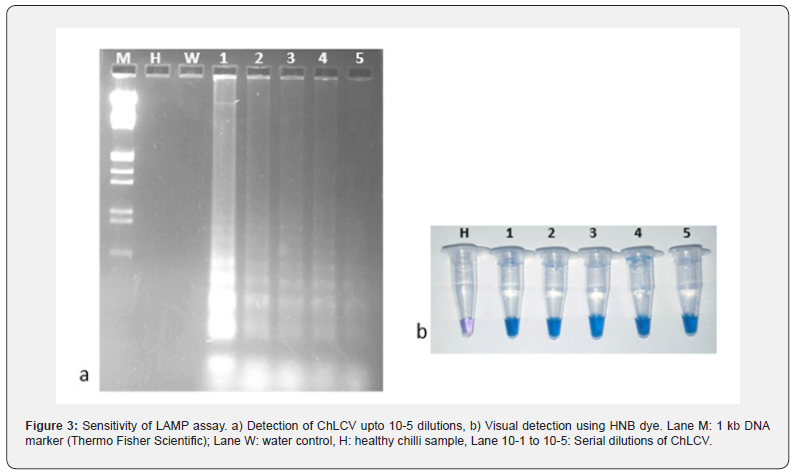
The sensitivity of assay was analyzed with the serial dilutions ranging from 10-1 to 10-5 of total nucleic acid extracted from ChLCV infected leaf sample. The results showed that positive amplification was observed upto 10-5 dilution in the infected samples and displayed a positive reactions with a ladder-like amplification pattern, which clearly shows the sensitivity of LAMP assay (Figure 3a) and the same was visualized using HNB dye (Figure 3b). The experiment repeated thrice with two replication of each dilution.
Validation of LAMP assay
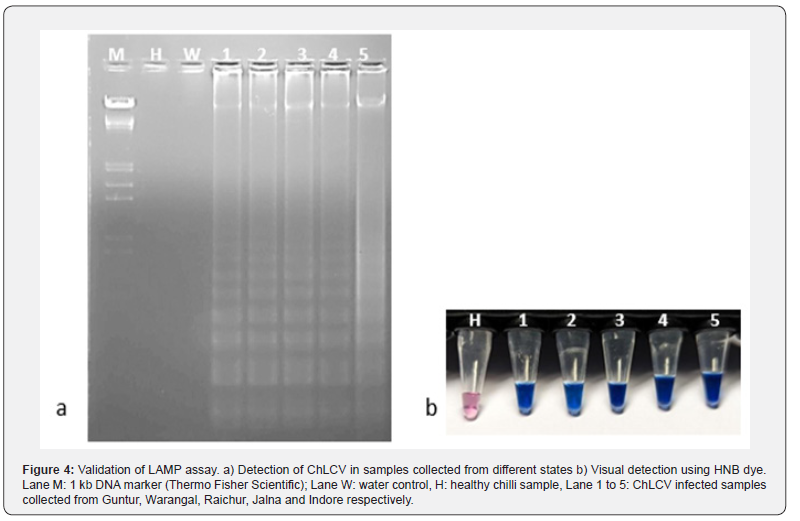
The LAMP assay was validated with the help of five leaf samples of chilli infected with ChLCV collected from different locations of India viz., Andhra Pradesh (Guntur, Warangal), Karnataka (Raichur), Madhya Pradesh (Lathur and Indore). The infected leaf samples initially subjected to specific begomovirus primers which showed positive with PCR amplicon of 1.2kb, then subjected to LAMP assay using LAMP primers. All the samples showed positive reactions with a ladder like pattern except the healthy and water control (Figure 4a). This was further confirmed by visualization of LAMP products by adding Hydroxy naphthol blue dye prior to PCR ampliciation, the positive samples showed sky blue color, whereas violet color was observed in healthy sample and water control (Figure 4b).
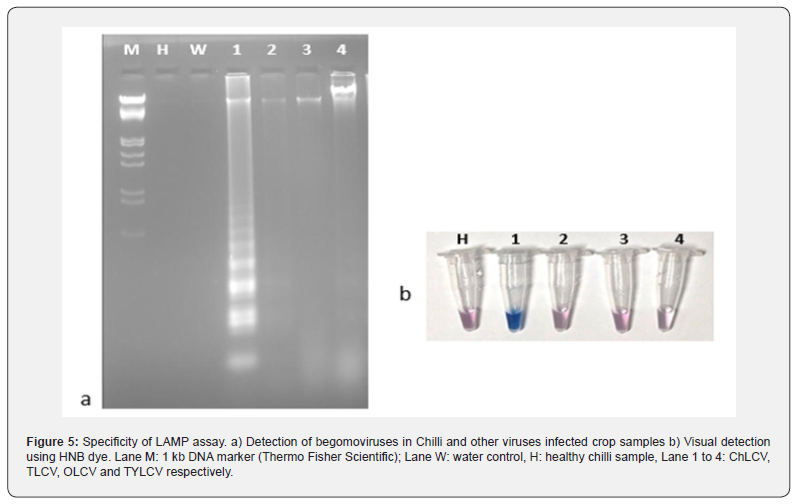
Specificity of LAMP assay
The specificity of LAMP assay was determined by performing LAMP assay using specific ChLCV LAMP primers with infected chilli samples and other begomoviruses infected samples such as tobacco (TbLCV), okra (OLCV) and tomato (ToLCV). The results showed that positive amplification was observed only in ChLCV infected samples and no amplification was seen in the samples infected with other viruses, healthy sample, and water control, which shows the specificity of LAMP assay to detect the ChLCV (Figure 5a). The experiment was repeated two times. Similar visual detection showed sky blue color in positive samples and violet like color in negative samples (Figure 5b).
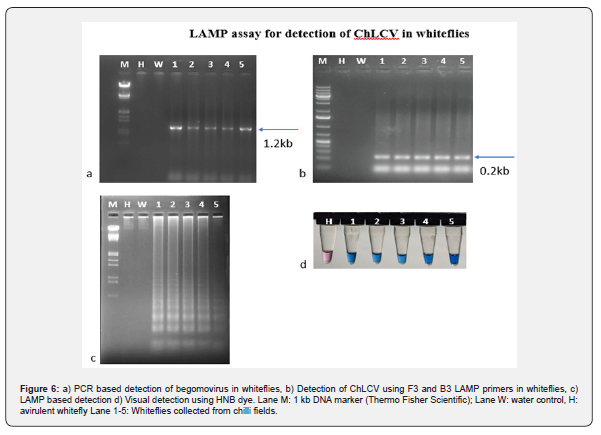
Detection of begomovirus in whiteflies using LAMP primers
Total nucleic acid was extracted from whiteflies were subjected to PCR amplification using LAMP primers of ChLCV-F3 and ChLCV-B3 (Figure 6a). The resulted PCR amplicon 180 bp was observed on agarose gel except the negative control (Figure 6b).
LAMP assay for detection of begomovirus in whiteflies
For the detection of ChLCV in the whiteflies collected from famers fields, the total genomic DNA extracted from whiteflies were subjected LAMP assay using ChLCV specific LAMP primers. The results showed that positive amplification of virus in whiteflies collected from field with a ladder like amplification pattern and there is no amplification was observed in healthy whitefly DNA and water control (Figure 6c). The same was observed with visual detection using HNB dye which showed sky blue color in positive samples and violet color in negative control (Figure 6d).
Discussion
Chilli is susceptible to numerous pathogens, particularly viruses, posing a significant threat for its production and resulting in substantial yield losses in worldwide [18]. Over sixty-five distinct viruses have been reported to infect chilli crop globally causing leaf curl disease in chilli and yield up to 100% losses in marketable fruits [2]. Of these ChiLCV is one of the most prevalent monopartite begomoviruses affecting solanaceous and non-solanaceous hosts, often in conjunction with various betasatellites [4]. Efficient management strategies for any disease hinge on the prioritized identification of the causal agent. Consequently, a prerequisite step involves in the development of diagnostic procedure is most efficient, stable, robust, rapid, and reliable, enabling the specific detection of plant pathogens. Although PCR using universal primers is the most commonly employed method for detecting begomoviruses in many crops, but it is unsuitable for field use due to lacking convenient portable instruments [19]. Furthermore, the overall time required for PCR detection, encompassing DNA extraction and electrophoresis, extends over several hours. As a result, there is a pressing need for a swift and uncomplicated detection assay to facilitate routine sampling of plant viruses [7].
In recent times, the LAMP assay has been documented for identifying various plant pathogens, such as viruses. This technique is asserted to be swift, remarkably sensitive, and more dependable than traditional PCR [13,20]. In the present LAMP assay was used for detection of ChLCV in chilli and its vector. The LAMP primers developed in the study was highly specific to ChLCV Guntur isolate. Similar techniques was applied for detection various begomoviruses infecting many crops and its whitefly vector [11- 13]. For the convenient implementation of the LAMP assay in the field, one can easily monitor the amplification process using visual examination without the need for specialized equipment. The amplification inspection involves observing a color transformation triggered by the introduction of specific fluorescent or metal dyes into the reaction mixture. HNB, functioning as a metal indicator for calcium and serving as a colorimetric reagent for alkaline earth metal ions, enables precise discrimination of a positive reaction, as evidenced by the shift in color from violet to sky blue [11,20]. In the present study, visual detection of LAMP products was assayed using hydroxy naphthol blue, which showed a color change of positive products to sky blue, while the control remained in violet color for non-positive samples. The detection of the virus was demonstrated by diluting the DNA to 10-5, showing the high sensitivity of the LAMP assay [6,10,11]. This assay was validated by testing other ChLCV infected samples collected from different states, which showed positive results, indicating high specificity. Additionally, the LAMP assay was used to detect the virus in whiteflies cryptic species collected from the chilli fields. Thus, in the present study, a standardised LAMP assay was developed for the detection of chilli leaf curl virus (ChLCV) in chilli and its vector in South India.
Acknowledgment
The authors are grateful to the Director, Indian Institute of Horticultural Research, Bangalore for providing research facilities and funding to conduct the research.
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Competing interests: All the authors declare that they have no competing interests.
Author contributions: BNT and V.V designed the experiment and wrote the manuscript, AC, RSJ, KMR, MM and MKR are associated with compilation of manuscript.
Availability of data and materials: Data will be made available upon request.
References
- Zehra SB, Ahmad A, Sharma A, Sofi S, Lateef A, et al. (2017) Chilli leaf curl virus an emerging threat to chilli in India. Int J Pure App Biosci 5(5): 404-414.
- Nigam K, Suhail S, Verma Y, Singh V, Gupta S (2015) Molecular characterization of begomovirus associated with leaf curl disease in chilli. World J Pharm Res 4(3): 1579-1592.
- Senanayake DMJB, Mandal B, Lodha S, Varma A (2007) First report of Chilli leaf curl virus affecting chilli in India. Plant Pathol 56(2).
- Shingote PR, Wasule DL, Parma VS, Holkar SK, Karkute SG, et al. (2022) An Overview of Chili Leaf Curl Disease: Molecular Mechanisms, Impact, Challenges, and Disease Management Strategies in Indian Subcontinent. Front Microbiol 13: 899512.
- Roy B, Dubey S, Ghosh A, Shukla SM, Mandal B, et al. (2021) Simulation of leaf curl disease dynamics in chili for strategic management options. Scientific Rep 11(1): 1010.
- Jeevalatha A, Kaundal P, Kumar R, Raigond B, Kumar R, et al. (2018) Optimized loop-mediated isothermal amplification assay for tomato leaf curl New Delhi virus-[potato] detection in potato leaves and tubers. Eur J Plant Pathol 150: 565-573.
- Wilisiani F, Tomiyama A, Katoh H, Hartono S, Neriya Y, et al. (2019) Development of a LAMP assay with a portable device for real-time detection of begomoviruses under field conditions. J Virol Methods 265: 71-76.
- Notomi T, Mori Y, Tomita N, Kanda H (2015) Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol 53(1): 1-5.
- Nagamine K, Hase T, Notomi TJMCP (2002) Accelerated reaction by loop-mediated isothermal amplification using loop primers. Molecular and cellular probes, 16(3): 223-229.
- Kuan CP, Wu MT, Lu YL, Huang HC (2010) Rapid detection of squash leaf curl virus by loop-mediated isothermal amplification. J Virol Methods 169(1): 61-65.
- Naganur P, Premchand U, Shankarappa KS, Mesta RK, Manjunatha C, et al. (2019) Development of a loop-mediated isothermal amplification assay for detection of tomato leaf curl New Delhi virus in ridge gourd [Luffa acutangula (L.) Roxb.]. Int J Curr Microbiol Appl Sci 8: 2282-2295.
- Venkataravanappa V, Ashwathappa KV, Reddy CNL, Shankarappa KS, Reddy MK (2020) Characterization of tomato leaf curl New Delhi virus associated with leaf curl and yellowing disease of Watermelon and development of LAMP assay for its detection. 3 Biotech 10(6): 1-12.
- Krishnan N, Kumari S, Kumar R, Pandey KK, Singh J (2022) Loop-mediated isothermal amplification assay for quicker detection of tomato leaf curl Joydebpur virus infection in chilli. J Virol Methods 302: 114474.
- Bellows TS, Perring TM, Gill RJ, Headrick DH (1994) Description of a species of Bemisia (Homoptera: Aleyrodidae). Ann Entomol Soc Am 87(2): 195-206.
- Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12(1): 13-15.
- Venkataravanappa V, Lakshminarayana Reddy CN, Jalali S, Reddy MK (2012) Molecular characterization of distinct bipartite begomovirus infecting bhendi (Abelmoschus esculentus L.) in India. Virus Genes 44(3): 522-535.
- Venkataravanappa V, Swarnalatha P, Reddy CNL, et al. (2016) Association of recombinant Chilli leaf curl virus with enation leaf curl disease of tomato: a new host for chilli begomovirus in India. Phytoparasitica 44(2): 213-223.
- Thakur H, Jindal SK, Sharma A, Dhaliwal MS (2018) Chilli leaf curl virus disease: a serious threat for chilli cultivation. J Plant Dis Prot 125: 239-249.
- Parida M, Sannarangaiah S, Dash PK, Rao PVL, Morita K (2008) Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol 18(6): 407-421.
- Panno S, Matić S, Tiberini A, Caruso AG, Bella P, et al. (2020) Loop mediated isothermal amplification: principles and applications in plant virology. Plants 9(4): 461.






























