Spinocerebellar Ataxia 8: Novel Mutation Identification by Whole Exome Sequencing in an Indian Subject
Ashish Dubey1, Priyanka Vishwakarma1*, Shashank Upadhyay2, Amit Joshi2, Deepika Kalo1 and Vishal Kumar Mishra3
1 Department of Clinical Genomics, Redcliffe Lifetech Private Limited, India
2 Department of Biotechnology, Invertis University, India
3 Department of Bioinformatics, Redcliffe Lifetech Private Limited, India
Submission: March 18, 2022; Published: April 05, 2022
*Corresponding author: Priyanka Vishwakarma, PhD, Department of Clinical Genomics, Redcliffe Lifetech Private Limited, H-55, 3RD Floor, Sector 63, Gautam Buddha Nagar, Noida, Uttar Pradesh 201301, India
How to cite this article: Ashish D, Priyanka V, Shashank U, Amit J, Deepika K, et al. Spinocerebellar Ataxia 8: Novel Mutation Identification by Whole Exome Sequencing in an Indian Subject. JOJ Case Stud. 2022; 13(3): 555863. DOI: 10.19080/JOJCS.2022.13.555863.
Abstract
Background: Spinocerebellar ataxia (SCA) is an advanced, deteriorating, genetic disease with the multiple types, each of which could be considered as neurological condition in its own right.
Case presentation: A novel variant in the SYNE1 gene: c.5635_5636insT, classified as likely pathogenic, was identified in a patient with a phenotype attuned with Ataxia, also with the complaints of the blurring vision, born in consanguineous marriage, history of vertigo, cerebellar atrophy and hypothyroidism with no positive family history.
Conclusion: This case is added to the Novel variant in SCA subtype 8 in Indian subcontinent it is novel case Identified. This case showed the utility of Whole exome sequencing and final validation by Sanger sequencing technique. This finding adds the utility of the current technologies in the undiagnosed cases of ataxia with negative family history.
Keywords: Spinocerebellar ataxia; Novel mutation; Neurogeneration; Likely pathogenic mutation; Ataxia
Abbreviations: HPO: Human Phenotype Oncology;dbSNP: Database Single Nucleotide Polymorphism; gnomAD: The Genome Aggregation Database; LRT: Likelihood Ratio Test; SIFT: Sorting Intolerant from Tolerant; OMIM: Online Mendelian Inheritance in Man; HGMD: Human Gene Mutation Database: SYNE:1 Spectrin repeat containing nuclear envelope 1gene; SCAR: Spinocerebellar Ataxia Recessive
Introduction
Spinocerebellar ataxia (SCA) is a word declaring to a group of the hereditary ataxias that are characterized by the deteriorating dissimilarities in the part of the brain linked to the movement control (cerebellum), and sometimes in the spinal cord region [1]. SCAs are a heterogeneous group of the neurodegenerative disorders and it is considered as the progressive cerebellar ataxias associated with a variable arrangement of the pyramidal and the extrapyramidal signs, such as neuropathy, ophthalmoplegia, cognitive impairment and the epileptic features [2]. Till date, more than 30 unlike SCAs have been demarcated and the causative mutations recognized in the more than 20 of them [3]. Most of the causative mutations are include the trinucleotide repeat expansions, which can be either translated or positioned in the introns and the untranslated 5’ or 3’ regions. The Indels and the missense mutations have also been designated [4]. The most frequent types are caused by the expansions in the coding CAG repeats. The occurrence of the SCA subtypes varies among populations [5].
Ataxia word describes a lack of the muscle synchronization in the course of voluntary activities typically due to the pathology in the cerebellum and its acquaintances, such as spinocerebellar and pontocerebellar pathways. Although it is a neurological symptom, there are the several inherited disorders that are due to this unstable movement of the gait and limbs, comprising Friedreich’s ataxia (FA), spinocerebellar ataxia (SCA), episodic ataxia (EA), ataxia telangiectasia (AT), and Machado–Joseph disease (MJD). Ataxias can signify as the sporadic or the hereditary and are allocated into the five key groupings: mitochondrial, metabolic, the defective DNA repair, abnormal protein folding and the degradation, and the channelopathies. On the basis of neurological examinations, ataxias can be mainly confidential into the pure form of cerebellar ataxias and those in which the extra neurological discrepancies are also existing [8].
The Autosomal recessive spinocerebellar ataxia type 8 (ARCA1/SCAR8) is caused by mutations of the SYNE1 gene. This disease was principally designated in the families from Quebec (Canada) with a phenotype of pure cerebellar syndrome, but in recent years has been reported with a more variable clinical phenotype in other countries. Cases have recently been described of muscular dystrophy, arthrogryposis, and cardiomyopathy due to SYNE1 mutations.
SCAR8 (OMIM#610743) has been first described in the families from Beauce and Bas-St-Laurent (Quebec, Canada). This disease is formed by the mutations in the spectrin repeat containing nuclear envelope 1 (SYNE1) gene, positioned on the chromosome 6 [5-7]. This gene covers 146 exons and it encodes a giant protein encompassing 8797 number of amino acids, known as nuclear envelope spectrin repeat protein 1 (nesprin 1). The protein is expressed in Purkinje cells, the olivary bodies, and in the myocytes; it has 4 domains (one presenting the spectrin-like structure characteristic of membrane-anchored proteins) and plays an important role in maintaining the structure of the cell, as it fixes the nuclear lamina to the cytoskeleton and contributes to the organisation of cytoplasmic organelles [4-6].
Clinical Summary
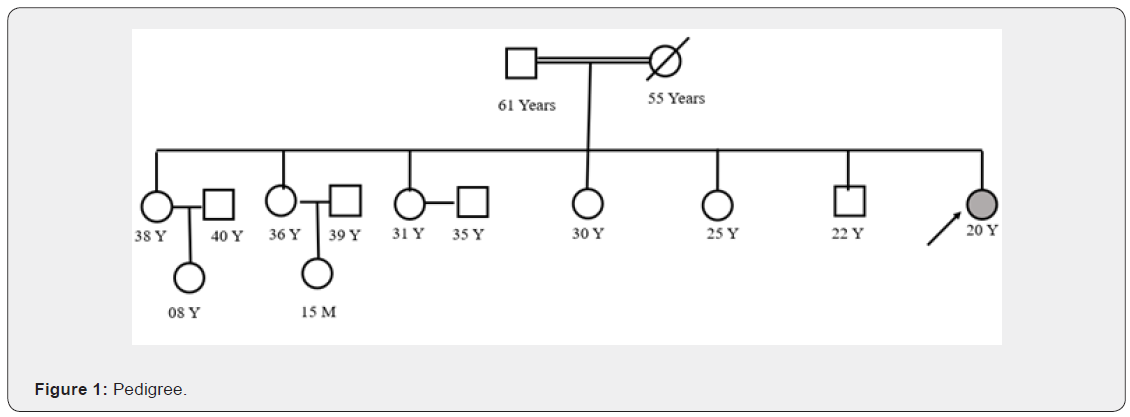
This patient is 20 years old, female had come to the clinician with the criticisms of the blurring vision, born in consanguineous marriage (Figure 1), history of vertigo, MRI brain suggested cerebellar atrophy and hypothyroidism, SCA repeat mutation panel testing has been found normal in this patient, also checked for the freidrich ataxia which was also found normal in this patient. No family history has been provided.
Materials and Methodology
The DNA extraction from blood was used to perform the targeted gene capture by using a custom capture kit. The libraries were sequenced to mean >100X coverage on Ilumina sequencing platform. The sequences obtained are aligned to human reference genome (GRCh37/hg19) and variant analysis was performed using set of different Bioinformatics Pipelines.
Variant analysis
Golden Helix VarSeq 2.2.0 is a clinical genomics interpretation and reporting platform from Golden Helix. The variant annotation engine includes algorithms to identify variant impact on gene using both public content (ClinVar, HPO, links to dbSNP, gnomAD and in-silico predictors - GERP++, PhyloP, PhyloP LRT, SIFT and PolyPhen2. VarSeq allows quick filtering and evaluation of variants. Clinically relevant variants were annotated using published variants in literature and a set of diseases databases – dbSNP, ClinVar, OMIM and HGMD. Common variants are filtered based on allele frequency in gnomAD. Only non- synonymous and splice site variants found in the clinical exome panel consisting of specific set of genes were used for clinical interpretation. Silent variations that do not result in any change in amino acid in the coding region are not reported.
Result
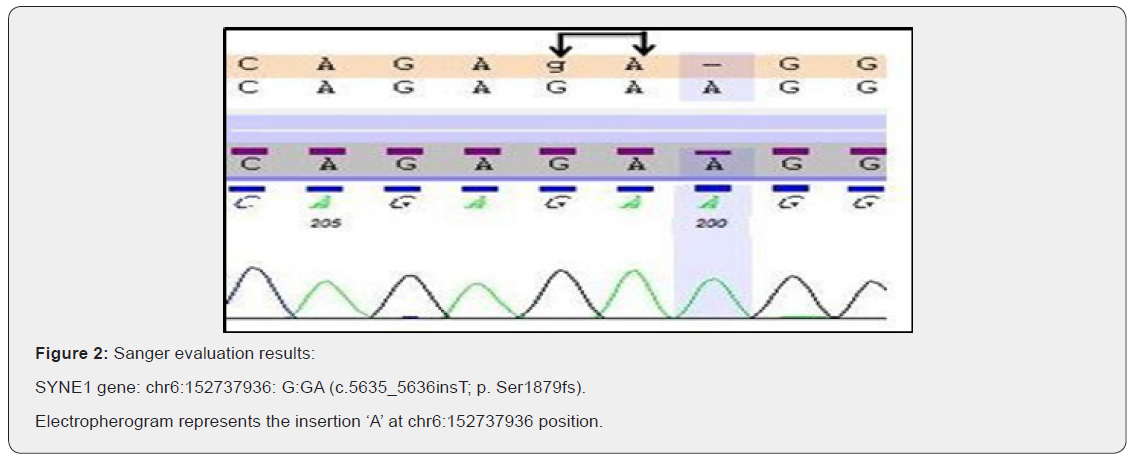
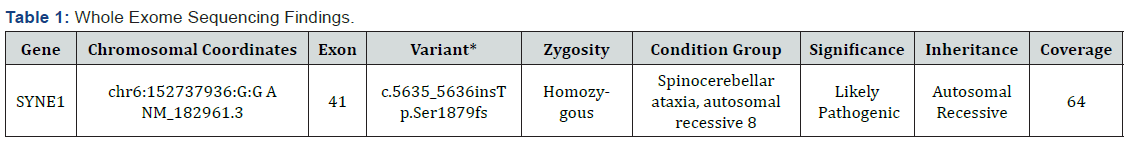
In this study, we found a novel variant (c.5635_5636insT p. Ser1879fs) in SYNE1 gene in exon 41 by the help of widely used technique whole exome sequencing and also this variant was successfully validated by the gold standard method Sanger sequencing (Figure 2). The individual carries two copies of a frameshift variant in SYNE1 gene which is predicted to cause a frameshift and consequent premature truncation of the protein. This variant seems to be novel as it has not been previously reported in literature. Since, this variant is predicted to produce a truncated protein which might result in loss-of-function, therefore, this variant has been labelled as likely pathogenic. This variant has been labelled as likely pathogenic (likely to disease causing) (Table 1) according to the ACMG (American College of Medical Genetics) guidelines [8].
Discussion
Next-generation sequencing technologies authorize a wide-ranging analysis of the entire genome and the exome and as such have affectedly progressed the field of the biological and the biomedical research, especially in the area of Mendelian diseases. Nevertheless, these new methodologies also have the numerous boundaries; for example, though the target coverage is continually improving day by day, on the other hand a complete coverage will probably never been reached since the specific genomic regions such as the GC-rich areas and the repetitive elements are very challenging to amplify and large copy number variations are very stiff to detect easily. Despite this, here we report on several movements disorder studies in which NGS technologies have successfully been employed for disease causing mutations’ identification [9]. Some of these findings have already been replicated in several studies reported from different subcontinents [10-12].
SCA subtype 8 is one type of Ataxia mid the group of inherited ailments of the central nervous system (CNS). As in additional inherited Ataxia, SCA8 is the result of the genetic flaws that leads to the diminishing of the particular nerve fibres carrying the messages to and from the brain, subsequent in degeneration of the cerebellum part of the brain (the harmonization centre of the brain part). SCA8 is a relatively rare form of Ataxia; its incidence is fewer than 1 out of 100,000. SCA type 8 (SCA8) is an inherited neurodegenerative disorder caused by a CTG trinucleotide repeat expansion in a noncoding gene of unknown function. SYNE1 gene Spinocerebellar ataxia, autosomal recessive 8 Autosomal recessive spinocerebellar ataxia-8 (SCAR8) is a slowly progressive neurodegenerative disorder characterized by gait ataxia and other cerebellar signs, such as nystagmus and dysarthria. The age at onset is highly variable, and but most often is in the second or third decades. Lots of study have been reported in different populations on different SCA subtypes. [13-17] in this study we firstly reporting a novel likely pathogenic variant in SCA subtype 8 in the Indian subcontinent.
The disorder was initially documented in the patients of French Canadian ancestry, most of whom have reported a comparatively 'pure' form of the disorder. However, the subsequent studies have shown that SCAR8 ensues worldwide and the most commonly manifests with additional features, including the spasticity, secondary musculoskeletal abnormalities, and ocular movement anomalies, consistent with a 'complicated' phenotype. Brain imaging typically shows cerebellar atrophy, sometimes with pontine involvement. Rare patients may have an early-onset multisystemic disorder with impaired intellectual development and respiratory dysfunction [OMIM 610743].
The SYNE1 gene encodes nesprin-1 protein, a member of the spectrin family of structural proteins that link with the nuclear plasma membrane to the actin cytoskeleton. The full-length protein is very large (1 MDa) and contains 8,797 amino acids; SYNE1 encodes multiple isoforms that show differential tissue expression [MIM 608441]. The SYNE1 gene provides the directions for making a protein called Syne-1 that is found in many tissues, but it appears to be especially critical in the brain region. The Syne-1 protein shows a very chief role in the preservation of the part of the brain that coordinates to the movement called cerebellum.
Conclusion
In conclusion this study showed the utility of Whole exome sequencing in the misdiagnosed pathogenic case. By this study we reported a novel pathogenic mutation in the Indian subcontinent. Whole exome approach provides a major advance in likely diagnostic yield in the suspected inherited mutation containing cases, this methodology may uncover unexpected findings such as mutations in genes known to cause familial. Next Generation Sequencing (NGS) technology overwhelms the constraint of single gene analysis and could deliver cheaper and more rapid large-scale DNA sequencing.
Authors Contribution
PV is working as Scientist, have made substantial contributions to conception and design, who worked and compiled the data and drafted this manuscript. AD did a substantial contribution in finalizing and critical review of the manuscript. SD is a Scientist who reviewed the study. DK is working as a Scientist who gave her valuable input or acquisition of data, or analysis and interpretation of data during the manuscript preparation. AJ is a scientist who reviewed the study thoroughly. VKM is bioinformatics Scientist, who provided his major input during data analysis. All authors contributed to the article and approved the submitted version.
Acknowledgement
The authors would like to thank all the study participants. We are grateful to the physicians who referred patients for the further investigation.
Compliance with Ethical Standards
Informed consent was obtained from all subjects involved in the study.
References
- Begley CM, Gyte GM, Devane D, McGuire W, Weeks A (2019) Active versus expectant management for women in the third stage of labour. Cochrane Database of Systematic Reviews 2(2): CD007412.
- Melamed N, Haroush AB, Chen R, Kaplan B, Yogev Y (2009) Intrapartum cervical lacerations: characteristics, risk factors, and effects on subsequent pregnancies. Am J Obstet Gynecol 200(4): 388.e1-388.e4.
- https://www.bettersafercare.vic.gov.au/clinical-guidance/maternity/postpartum-haemorrhage-pph-prevention-assessment-and-management#goto-risk-factors
- Landy HJ, Laughon SK, Bailit JL, Kominiarek MA, Quintero VHG, et al. (2011) Characteristics Associated with Severe Perineal and Cervical Lacerations During Vaginal Delivery. Obstet Gynecol 117(3): 627-635.
- Parikh R, Brotzman S, Anasti JN (2007) Cervical lacerations: some surprising facts. Am J Obstet Gynecol 196(5): e17-e18.
- Djokovic D, Costa C, Martins A, Abushad S (2015) Spontaneous delivery through a cervical tear without cervical os dilation. Clin Case Rep 3(1): 3-6.
- M Widmer, G Piaggio, GJ Hofmeyr, G Carroli, A Coomarasamy, et al. (2020) Maternal characteristics and causes associated with refractory postpartum haemorrhage after vaginal birth: a secondary analysis of the WHO CHAMPION trial data. BJOG 127(5): 628-634.
- Kellie FJ, Wandabwa JN, Mousa HA, Weeks AD (2020) Mechanical and surgical interventions for treating primary postpartum haemorrhage. Cochrane Database Syst Rev 7(7): CD013663.






























