Clinical Exome Sequencing in Cases with Autosomal Recessive Cerebellar Ataxia: A Case Control Study
Priyanka Vishwakarma, Ambreen Asim, Sarita Agarwal* and Kausik Mandal
Department of Medical Genetics, Sanjay Gandhi Postgraduate Institute of Medical Sciences (SGPGIMS), India
Submission: February 28, 2022; Published: March 10, 2022
*Corresponding author: Sarita Agarwal PhD, Department of Medical Genetics, Sanjay Gandhi Postgraduate Institute of Medical Sciences (SGPGIMS), Lucknow 226014, India
How to cite this article: Priyanka V, Ambreen A, Sarita A, Kausik M. Clinical Exome Sequencing in Cases with Autosomal Recessive Cerebellar Ataxia: A Case Control Study. JOJ Case Stud. 2022; 13(2): 555860. DOI: 10.19080/JOJCS.2022.13.555860.
Abstract
Autosomal recessive cerebellar ataxias (ARCA) are a rare group of neurodegenerative disorders caused by the deterioration of the cerebellum, brainstem, and spinal cord. Clinical exome sequencing (CES) has successfully detected various forms of SCA in patients presenting with overlapping features in the past and therefore, we have employed CES in 10 ARCA cases of Indian origin with clinical features, such as, progressive ataxia, Nystagmus, dysarthria, tremor. Peripheral blood in EDTA vial was collected after obtaining written informed consent. Cases underwent series of biochemical test screening and brain MRI was conducted. Genomic DNA was extracted followed by Clinical Exome Sequencing (CES). The results were confirmed by Sanger sequencing. CES identified a novel homozygous pathogenic variant, c.5365del (p. Glu1789SerfsTer25), in the exon 11 of SETX gene, in a 21-year female proband born into a consanguineous family and diagnosed with Spinocerebellar ataxia-1 (SCA-1) with Autosomal oculomotor Apraxia type 2 (AOA2). Laboratory test showed an elevated alpha-fetoprotein (AFP) level. CES successfully confirms the identification of novel homozygous deletion mutation in SETX gene located on exon 11 in 21-year female proband diagnosed with AOA2.
Keywords: Autosomal recessive; Cerebellar ataxia; Ataxia; Repeat expansion; Clinical exome sequencing
Introduction
Boder and Sedgwick first time observed the atypical eye movements in the cerebellar ataxia associated with oculomotor apraxia (OA) in Ataxia Telangiectasia (AT) patients [1]. OA is further classified into ataxia with oculomotor apraxia type 1(AOA1), ataxia with oculomotor apraxia type 2 (AOA2), AT and AT-like disorders. All above mentioned disorders are inherited in autosomal recessive fashion and have a characteristic feature of atypical eye movement. Other features include, cerebellar gait and limb ataxia along with neuropathy and cerebellar degradation. All these subtypes are also characterised by elevated alpha-fetoprotein levels in the serum (except for Ataxia Telangiectasia like disorders). Beside the clinical and biochemical parameters, these sub-types also share a common underlying pathophysiological mechanism. The causative genes for the occurrence of these ataxia encodes the proteins that are involved in the single-stranded and double stranded break repair mechanism [2].
Out of all the above-mentioned ataxia associated oculomotor apraxia sub-types, highest prevalence has been recorded for Ataxia Telangiectasia that accounts for 1/300,000 and is caused by the pathogenic variants in ATM gene that affects the function of encoding protein known as, phosphatidylinositol-3 kinase protein [2]. These patients are more vulnerable for other types of immunodeficiencies and cancer predisposition. While, Ataxia Telangiectasia like disorders is the rare form of disorder and is caused due to the pathogenic mutation arising in the human meiotic recombination 11 genes and the symptoms are relatively milder in nature compared to Ataxia Telangiectasia [3,4]. AOA1 is early onset with neuropathy, mental retardation and is often characterized by reduced level of albumen and elevated level of cholesterol level in the serum. The causative gene is APTX that encoded for aprataxin protein that is essential in the single-stranded DNA break repair mechanism [2].
AOA2 is adolescent-onset type and has an estimated prevalence of approximately 1 in 900,000. It is characterized by atrophy of cerebellum vermis that predominantly affects the gait, sensory polyneuropathy. Elevated levels of alpha-fetoproteins have also been reported in this sub-type. Approximately, fifty percent of these cases are associated with oculomotor apraxia. It is caused by SETX gene mutation that is located on chromosome 9 and has 24 exons. This gene encodes a 2,677 amino acid long Senataxin protein [5,6].
Senataxin is a helicase protein that function primarily to protects DNA from the damage and also contributes in the other important cellular functions like transcription regulation, posttranscriptional modification of mRNA and meiotic [5,7,8].
Pathogenic mutations in the SETX gene are previously associated with juvenile form of amyotrophic lateral sclerosis type 4 (ALS 4), autosomal dominant proximal spinal muscular atrophy, and in the tremor-ataxia syndrome (TAS). ALS 4 is autosomal dominant in nature, is characterized by wasting of muscles and lacks the participation of the sensory system [9-16]. Patients suffering from TAS are characterized by tremors and absence of peripheral neuropathy. On the contrary, autosomal dominant proximal spinal muscular atrophy is characterized with muscle wasting and elevated levels of creatine kinase in the serum [2].
Various types of mutations have been identified leading to AOA2 in the past ranging from small insertion/deletion to frameshift mutations [12-17]. However, the profile of SETX gene mutations is thought to be more polymorphic and is considered to be hotspot for novel variants, thus making a difficult task for a clinician to diagnose those patients having overlapping phenotypes with other forms of ataxias including AOA1, AOA2, ataxia telangiectasis and ataxia telangiectasis like disorder. Previous studies have shown that clinical exome sequencing has successfully detected various forms of SCA in patients presenting with overlapping features [14].
Thus, the current study highlights the use of clinical exome sequencing in ten cases with autosomal recessive cerebellar ataxia with clinical features, such as, progressive ataxia, Nystagmus, dysarthria, tremor.
Study Subjects
We recruited ten ARCA patients from the OPD of Department of Medical Genetics, Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow. Informed consent from each participant was taken and the study approved by institutional ethical committee (2017-20-PhD-95PGI/BE/259/2017).
The study subjects had following clinical features, such as, progressive ataxia, Nystagmus, dysarthria, tremor. These study subjects showed autosomal recessive inheritance pattern. In all subjects MRI of the head and cervical cord; and electromyography (EMG) was performed. Series of laboratory examinations were conducted in the study subjects, such as, the complete blood cell count, ESR, thyroid hormone profiling and serum evaluation of C - reactive protein, alpha fetoprotein, vitamin B6 and vitamin B12 level.
All subjects who experienced an early onset of ataxia-related features (unlikely to be found in most of the SCA cases), having family pedigree with recessive inheritance pattern and were found to be negative for FRDA mutations were included in the study. The subjects with FRDA expansion mutation were excluded from the study. All the study subjects underwent clinical exome sequencing.
4.1. DNA isolation
Genomic DNA was extracted from Qiagen kit according to manufacturer instructions. We have excluded the GAA repeats present in the FXN gene causing Fredrich Ataxia (FRDA) as reported by Muthuswamy & Agarwal 2013 [17]. All ten samples recruited for CES (Ataxia panel) were found to be negative for GAA repeat expansion leading to FRDA.
4.2. Clinical exome sequencing
Targeted gene capture (Illumina sequencing platform) using a custom capture kit was used to sequence the entire protein-coding regions known as exons of the genome. The genomic libraries were prepared and then sequencing was done to give mean >80-100 X coverage. The read obtained were then aligned to human reference genome (GRCh37/hg19).
Variant annotation of clinically relevant mutations was done by using the extensive literature search of published variants and, also, a search was made on various databases such as, ClinVar, OMIM, HGMD and SwissVar [8-11]. Based on allele frequency, variant filtering was done in databases such as 1000 Genome Phase 3, ExAC, gnomAD and dbSNP (v151) [12-16]. The pathogenic variants that were non-synonymous were identified by bioinformatic tools such as, PolyPhen-2, SIFT and MutationTaster2. The clinical exome panel comprise of 8332 genes having non-synonymous and splice-site variants. Reference sequence, ENST00000224140, was used for sequence alignment using online available bioinformatic softwares. Sanger sequencing was used to validate the identified variants.
Results
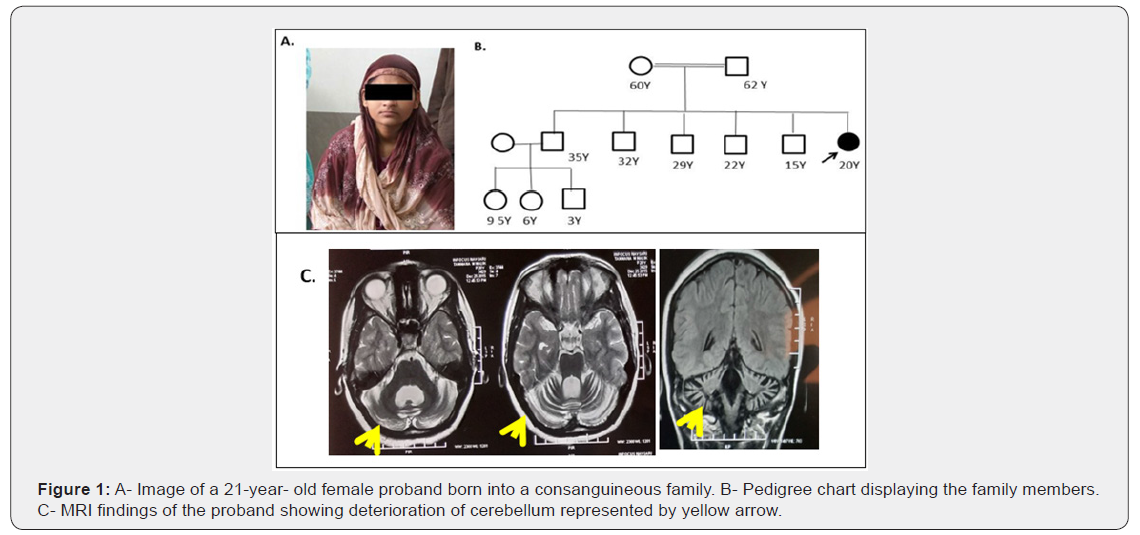
We identified a novel homozygous mutation in SETX gene exon 11 (c.5365del; p. Glu1789SerfsTer25) in one of the 21-year-old female proband belonging to a consanguineous family of Indian origin (Figure 1). The exon 11 of SETX gene was amplified and amplified products was subjected to Sanger sequencing to validate the results.
Discussion
A 21-years old female proband born from consanguineous family of Indian origin presented with classical features of juvenile form of ARCA. The age of onset of proband was 18 years and was initially diagnosed with cerebellar ataxia presented with unsteady walk and dysarthria (Figure 1). The phenotypic features of the proband agreed with the features of AOA2 with the following clinical features, such as, oculomotor apraxia, sensorimotor neuropathy and marked cerebellar atrophy. Electromyography (EMG) tests in the proband confirmed peripheral nerve damage. Elevated levels of AFP in the serum were also reported with cerebellar atrophy in the proband (Figure 1C). Various biochemical parameter screening of proband was also done including, CBC; profiling of thyroid hormone, ESR, C-reactive protein, and levels of Vitamin B6 and B12. All these parameters were found to be within the normal range in the proband. Figure 1 A & 1B shows the image and pedigree chart of a proband born into a consanguineous family. All the three siblings of the proband were normal.
The OA is associated with AOA2 [18- 22]. In the present study, the proband’s ocular movements were found to be normal further highlighting that OA may not influence the diagnosis of AOA2. Instead, due to early-onset of the disease, OA has not still appeared in the proband and therefore, this particular study requires a consequent follow-up.
Clinical exome sequencing identified the variant as a frameshift mutation having homozygous deletion of one bp in exon 11 of the SETX gene that resulted in premature truncation at protein position 25 amino acids downstream in the codon 1789. Figure 2A shows the an electropherogram showing a frameshift mutation on exon 11 of the SETX gene (chr9: g.135187155delC; c.5365del; p. Glu1789SerfsTer25) along with the alignment to the reference sequence. Figure 2B shows the graphic diagram of the SETX gene highlighting the distribution and location of exons along with the list of variants present on SETX gene reported till date.

Till date, approximately 125 variants have been reported in the SETX gene is associated with AOA2. However, the reduced prevalence of AOA2 was reportedly globally and sporadic in Asian population [7]. Among the mutations detected in SETX gene, the mutational hotspots are reported to be in exon 10 of SETX gene[2].On the contrary, our study has first time identified novel mutation in exon 11 of SETX gene in a proband of Indian origin. The mutations associated with AOA2 are supposed to interrupt the helicase activity of Senataxin leading to the occurrence of AOA2. However, few reports have highlighted the impairment of DNA repair processes finally causing cell death in the region of brain including the cerebellum, thereby leading to movement disabilities along with AOA2 [9].
The novel mutation reported here was predicted to be pathogenic by bioinformatics softwares and also reported pathogenic according to the AGMG guidelines [23], however, the findings need to be yet confirmed through functional studies [23]. Probably, the variant has hampered one of the important mechanisms of DNA repair in Senataxin causing AOA2 [15,24]. However, further functional investigations will be required to elucidate the pathogenicity of the identified novel mutations in the future.
AOA2 being a sporadic event, the early-onset diagnosis is considered to be a tough task for clinicians. Clinical features including, early-onset ataxia, oculomotor impairment, and autosomal recessive inheritance pattern, it is also necessary to monitor the levels of serum AFP. Along with these, EMG test on patients will also be beneficial to determine the atrophy of cerebellar. Elevated AFP can be a clear indication of AOA2 along with the EMG findings showing peripheral nerve damage. Then, in this scenario, SETX genemutations screening in these patients will be an important parameter to further confirm the diagnosis.
Conclusion
The current study confirms the utility of clinical exome sequencing in the identification a novel homozygous deletion mutation SETX gene on exon 11 in an individual with AOA2 clinical features. To the best of our knowledge, the present is the first report of a novel mutation in the SETX gene implicated in AOA2 disease from India.
Acknowledgment
Authors are greatful to Sanjay Gandhi Postgraduate Institute of Medical Sciences (SGPGIMS), Lucknow, Uttar Pradesh, India for providing the infrastructure and lab facilities for research work.
Funding
We are thankful to the Science and Engineering Research Board-Department of Science and Technology (SERB-DST, EMR/2016/007407), Government of India.
Author’s Contribution
Priyanka Vishwakarma is a research scholar has carried out the experiment and sample collection. Ambreen Asim, is working as a research associate, who helped in the critical analysis and preparation of the manuscript. Sarita Agarwal is a professor in the department of medical genetics, helped in shaping the research plan and in the critical analysis of the manuscript. Kausik Mandal, is working as a consultant, who provided the referrals as study samples.
Ethics Approval and Consent to Participate
The study was approved by Institution al ethical committee (2017-20-PhD-95PGI/ BE/259/2017) of Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow. All participated agreed to give written consent for the study.
References
- Boder E, Sedgwick RP (1958) Ataxia-telangiectasia: a familial syndrome of progressive cerebellar ataxia, oculocutaneous telangiectasia and frequent pulmonary infection. Pediatrics 21(4): 526-554.
- Nanetti L, Cavalieri S, Pensato V, Erbetta A, Pareyson D, et al. (2013)SETX mutations are a frequent genetic cause of juvenile and adult onset cerebellar ataxia with neuropathy and elevated serum alpha-fetoprotein. Orphanet J Rare Dis 8: 123.
- Vermeer S, van de Warrenburg BPC, Willemsen MAAP, Cluitmans M, Scheffer H, et al. (2011) Autosomal recessive cerebellar ataxias: the current state of affairs. J Med Genet 48(10): 651-659.
- Fogel BL, Lee JY, Lane J, Wahnich A, Chan S, et al. (2012) Mutations in rare ataxia genes are uncommon causes of sporadic cerebellar ataxia. Mov Disord 27(3): 442-446.
- Date H, Igarashi S, Sano Y, Takahashi T, Takahashi T, et al. (2004) The FHA domain of aprataxin interacts with the C-terminal region of XRCC1. Biochem Biophys Res Commun 325(4): 1279-1285.
- Bernard V, Minnerop M, Bürk K, Kreuz F (2018) Exon deletions and intragenic insertions are not rare in ataxia with oculomotor apraxia 2. BMC Med Genet 10: 87.
- Lu C, Zheng YC, Dong Y, Li HF (2016) Identification of novel senataxin mutations in Chinese patients with autosomal recessive cerebellar ataxias by targeted next-generation sequencing. BMC Neurol 16(1): 179.
- Bennett CL, La Spada AR (2015) Unwinding the role of senataxin in neurodegeneration. Discov Med 19(103): 127-136.
- Rothblum Oviatt C, Wright J, Lefton Greif MA, McGrath Morrow SA, Crawford TO, et al. (2016) Ataxia telangiectasia: a review. Orphanet J Rare Dis 11(1): 159.
- Moreira MC, Klur S, Watanabe M, Nemeth AH, Le Ber I, et al. (2004) Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet 36(3): 225-227.
- Suraweera A, Becherel OJ, Chen P, Rundle N, Woods R, et al. (2007) Senataxin, defective in ataxia oculomotor apraxia type 2, is involved in the defense against oxidative DNA damage. J Cell Biol 177(6): 969-979.
- Kayal AK, Goswami M, Das M, Masaraf H (2011) A case of Spinocerebellar Ataxia from ethnic tribe of Assam. Ann Indian Acad Neurol 14(2): 122-123.
- Airoldi G, Guidarelli A, Cantoni O, Panzeri C, Vantaggiato C, et al. (2010) Characterization of two novel SETX mutations in AOA2 patients reveals aspects of the pathophysiological role of senataxin. Neurogenetics 11(1): 91-100.
- Becherel OJ, Yeo AJ, Stellati A, Heng EYH, Luff J, et al. (2013) Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet 9(4): e1003435.
- Arning L, Epplen JT, Rahikkala E, Hendrich C, Ludolph AC, et al. (2013) The SETX missense variation spectrum as evaluated in patients with ALS4-like motor neuron diseases. Neurogenetics 14(1): 53-61.
- Bennett CL, La Spada AR (2015) Unwinding the role of senataxin in neurodegeneration. Discov Med 19(103): 127-136.
- Muthuswamy S, Agarwal S, Dalal A (2013) Diagnosis and genetic counseling for Friedreich's Ataxia: A time for consideration of TP-PCR in an Indian Setup. Hippokratia 17(1): 38-41.
- Li H (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5): 589-595.
- McLaren W (2010) Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26(16): 2069-2070.
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, et al. (2018) Ensembl. Nucleic Acids Res 46(D1): D754-D761.
- Plagnol V (2012) A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics 28(21): 2747-2754.
- Landrum MJ (2015) ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44(D1): D862-D868.
- Mutation Taster2.
- Orban P, Devon RS, Hayden MR, Leavitt BR (2007) Chapter 15 Juvenile amyotrophic lateral sclerosis. Handbook of Clinical Neurology 82: 301-312.






























