The Impact of Area Size and Fabric Type on Touch DNA Collected from Fabric
Salem K Alketbi1,2* and Goodwin W1
1University of Central Lancashire, UK
2General Department of Forensic Science and Criminology, UAE
Submission:May 03, 2022;Published:May 16, 2022
*Corresponding author:Salem K Alketbi, General Department of Forensic Science and Criminology, University of Central Lancashire, Preston, UK, Dubai Police, UAE
How to cite this article:Salem K Alketbi, Goodwin W. The Impact of Area Size and Fabric Type on Touch DNA Collected from Fabric. J Forensic Sci & Criminal Inves. 2022; 16(1): 555926 DOI:10.19080/JFSCI.2022.16.555926.
Abstract
Trace DNA is commonly found at crime scenes and is often collected from the victim’s clothing in cases of sexual assault, homicide, and theft, typically by tape lifting. This study investigated the influence of area size and fabric type on the Touch DNA collected from fabric. The amount of collected Touch DNA from the fabric was significantly affected by fabric type (p < 0.05) and the interaction between fabric size and collection type (p < 0.05). More trace DNA was recovered from fabric containing a high percentage of polyester than 100% woven cotton. Minitapes were very effective for sampling Touch DNA from small areas of fabric but equally effective to cotton swabs for larger areas.
Keywords: Forensic science; Trace DNA; Touch DNA; DNA recovery; SceneSafe fast minitape; Cotton swab; PrepFiler express BTA; AutoMate express; Quantifiler™ human DNA quantification kit; GlobalFiler™ PCR amplification kit
Abbreviations: DNA: Deoxyribonucleic Acid; UV: Ultraviolet Radiation; CS: Cotton Swab; NS: Nylon Swab; MT: Mini Tapes
Introduction
Trace DNA is commonly found at crime scenes and frequently collected from a wide range of items such as tools, weapons, clothes, or even consumed food to link suspects to their crimes [1-3]. It is a valuable type of evidence but more challenging to collect compared to other biological samples because the sampling can be affected by surface type [4], environmental factors [5,6], collection methods [2,4], collection techniques [7,8] and extraction methods [4]. In cases of sexual assault, homicide, and theft, the victim’s clothing is often sampled for trace DNA, typically by tape lifting [4,9-11], but other collection methods can also be useful for different fabrics [12]. Therefore, this study investigated the influence of area size and fabric type on Touch DNA collected from fabric.
Materials & Methods
Experimental setup and deposition
The fabrics tested were composed of 65% polyester and 35% cotton (FB1), a popular synthetic material used in the fashion industry, and 100% woven cotton (FB2), the second most popular material used in the fashion industry [13]. The fabric samples were cut into 5x7cm pieces for DNA deposition and collection (Figure 1). For the area size experiment, only the polyester/cotton fabric was cut into two sizes (SZ1 = 5x7cm and SZ2 = 10x 14cm) (Figure 2). For the DNA deposition process, a participant previously identified as high shedder was instructed to wash both hands with antibacterial soap, stop any activity for 10 minutes, then charge both hands with eccrine sweat by touching their forehead to load them with epithelial cells. The participant was then instructed to rub the fabric sample for 1min between both hands. This procedure was repeated for each deposition. The fabric samples were washed at 50°C, dried and sterilised before use with ultraviolet radiation (UV) for 25 minutes.
DNA recovery and extraction
SceneSafe Fast™ minitape (K545) (MT) and a Copan cotton swab (150C) (CS) were used to collect the DNA for the fabric size experiment, whereas only MT was used to collect the DNA for the fabric type experiment. Before collection, the CS was moistened with 100μL of sterile distilled water applied using a plastic spray bottle [7]. No water was added to the MT but to increase the amount of Touch DNA collected, each minitape was applied 16 times to the area [4]. Samples collected with CS and MT were cut directly into the tubes for extraction using PrepFiler Express BTA™ kit extraction protocol with AutoMate Express (using 460μL of lysis buffer instead 230μL) (EXT1) according to the manufacturer’s instructions. Full swab heads were used for CS and NS, and the lower sticky part of the minitape, with a final elution volume of 50μL.
DNA quantification, amplification, and analysis
Extracted samples were quantified using the Quantifiler® Trio DNA Quantification Kit, QuantStudio 5 Real-Time PCR (qPCR) and HID Real-Time PCR analysis software v1.3 (Thermo Fisher Scientific) according to the manufacturer’s instructions. DNA amplification was performed using the GlobalFiler™ PCR amplification Kit on an ABI GeneAmp® 9700 PCR System (Life Technologies) for 30 cycles. The amplified products were sizeseparated and detected on an ABI 3500 Genetic Analyzer (Life Technologies) using 1μl PCR product, 9.6μl Hi-Di™ formamide, and 0.4μl GeneScan™ 600 LIZ® Size Standard v2.0 (Thermo Fisher Scientific). Finally, statistical analysis was performed with RStudio using factorial analysis of variance (ANOVA) and Microsoft Excel. Blank samples were taken from the fabrics after sterilisation, and negative controls for the collection, extraction, and amplification process, all of which were negative for DNA when analysed.
Results & Discussion
Fabric type
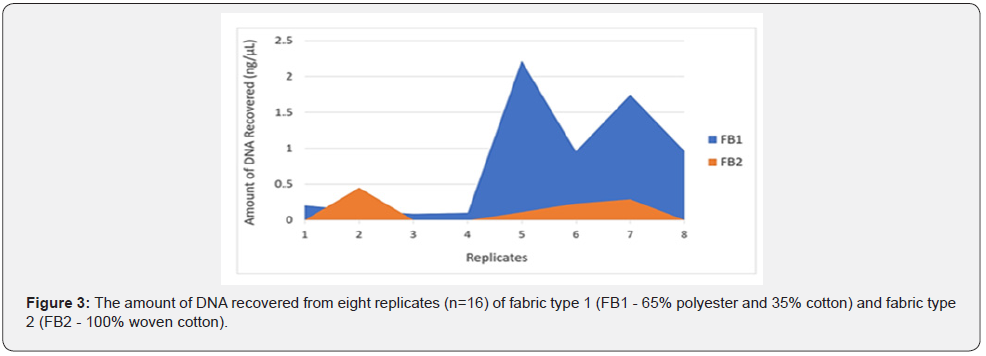
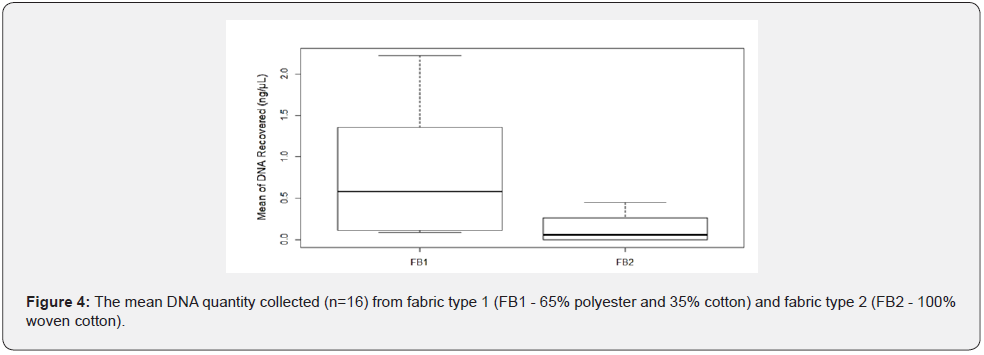
The amount of Touch DNA collected from the fabric was significantly affected by fabric type (p < 0.05), with more DNA recovered from FB1 composed of 65% polyester and 35% cotton than from FB2 composed of 100% woven cotton (mean: FB1 – 0.80 ng/μl vs. FB2-0.14ng/μL) (Figure 3 & 4). Some Touch samples collected from FB1 and FB2 were amplified to validate the DNA quality, with all samples producing full STR profiles but with some variation in the peak height between the fabric types. Samples collected from FB1 had a relatively higher peak height (RFU) compared to profiles from FB2.
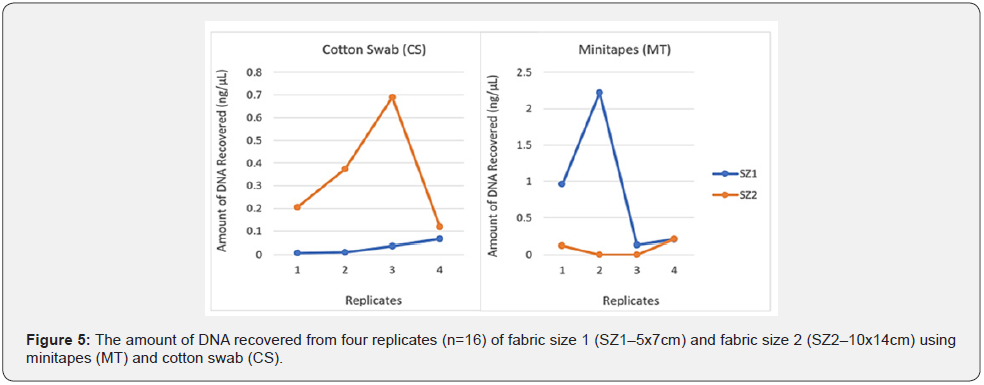
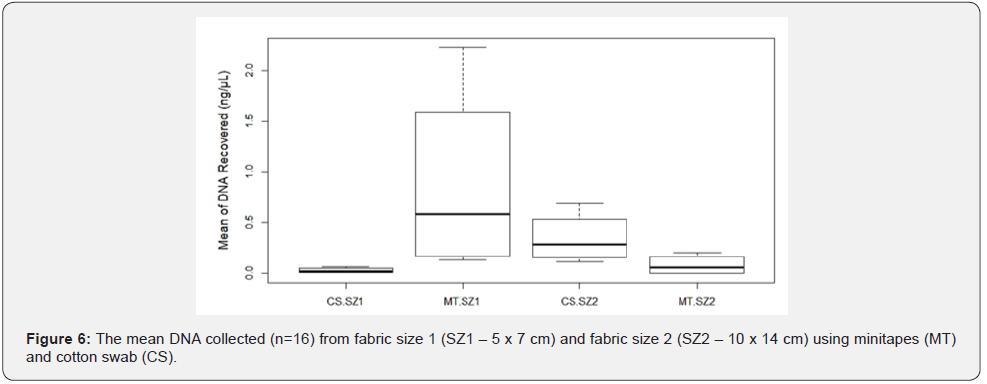
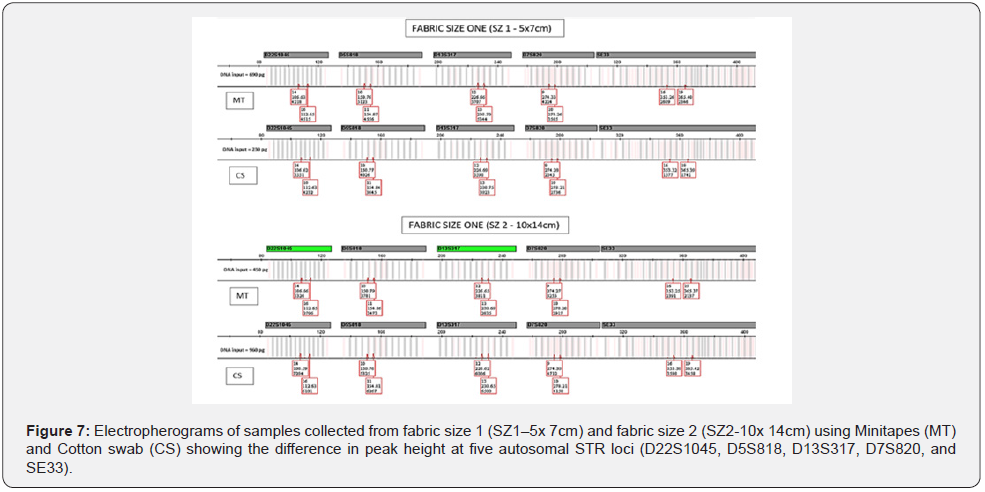
Fabric sizeand: The amount of Touch DNA collected from the fabric was significantly affected by the interaction between fabric size and collection type (p < 0.05), with more DNA recovered from fabric size 1 (SZ1 - 5 x 7cm) using minitapes (MT) than cotton swab (CS) (mean: MT-0.88ng/μL vs. CS-0.03ng/μL). However, with fabric size 2 (SZ2-10 x 14cm), more DNA was recovered using CS than MT (mean: MT-0.08ng/μL vs. CS-0.35ng/μL) (Figure 5 & 6). Minitapes are still very effective for collecting Touch DNA from fabric but are limited by the number of lifts which can be affected by sampling area size [11]. DNA collected from SZ1 and SZ2 was amplified to validate the sample quality, with all samples producing full STR Profiles but with some variation in the peak height (RFU) between the samples collected by each collection method for each fabric size (Figure 7). Samples collected by the MT from SZ1 had a relatively higher mean RFU than CS and it was the other way round for samples collected from SZ2.
Conclusion
Fabric type and the sampling area size can influence the amount of Touch DNA collected from clothing, with more trace DNA recovered from fabric composed of a high percentage of polyester than fabric composed of 100% woven cotton. Minitapes are very effective for sampling Touch DNA from small areas of fabric, but cotton swabs are equally effective for sampling larger areas of fabric.
Acknowledgement
This study was approved by the General Department of Forensic Science and Criminology in Dubai Police and ethical approval was granted by the School of Forensic and Applied Sciences, and the University of Central Lancashire’s Research Ethics Committee (ref. no. STEMH 912).
References
- Alketbi SK (2018) The affecting factors of Touch DNA. Journal of Forensic Research 9: 424.
- Verdon TJ, Mitchell RJ, Oorschot RA (2014) Swabs as DNA collection devices for sampling different biological materials from different substrates. Journal of Forensic Science 59(4): 1080-1089.
- Alketbi SK (2020) Collection of Touch DNA from rotten banana skin. International Journal of Forensic Sciences 5(4): 000204.
- Alketbi SK, Goodwin W (2019) The effect of surface type, collection, and extraction methods on Touch DNA. Forensic Science International. Genetics Supplement Series 7(1): 704-706.
- Alketbi SK, Goodwin W (2019) The effect of time and environmental conditions on Touch DNA. Forensic Science International. Genetics Supplement Series 7(1): 701-703.
- Alketbi SK, Goodwin W (2019) The effect of sandy surfaces on Touch DNA. Journal of Forensic, Legal & Investigative Sciences 5: 034.
- Alketbi SK, Goodwin W (2019) Validating Touch DNA collection techniques using cotton swabs. Journal of Forensic Research 10: 445.
- Alketbi SK, Goodwin W (2021) Touch DNA collection techniques for non-porous surfaces using cotton and nylon swabs. Journal of Scientific & Technical Research 36(3): 28608-28612.
- Hansson O (2009) Trace DNA collection-Performance of minitape and three different swabs, Forensic Science International. Genetics Supplement Series 2(1): 189-190.
- Stoop B (2017) Touch DNA sampling SceneSafe FastTM minitapes. Legal Medicine 29: 68-71.
- Alketbi SK (2022) The Impact of Collection Method on Touch DNA Collected from Fabric. Journal of forensic sciences & criminal investigation 15(5): 555922.
- Verdon TJ, Mitchell RJ, van Oorschot RA (2013) Evaluation of tape lifting as a collection method for Touch DNA. Forensic Science International: Genetics 8(1): 179-186.
- Textile Exchange (2017) Preferred Fiber Material Market Report.






























