Implications of Male Amelogenin Dropouts in Forensics
Anand Kumar*
DNA Division, State Forensic Science Laboratory, India
Submission:February 06, 2021;Published:February 22, 2021
*Corresponding author:Anand Kumar, DNA Division, State Forensic Science Laboratory, Rajasthan, 302016, (INDIA)
How to cite this article:Kertzman S, Kagan A, Vainder M, Hegedish O, Lapidus R, Weizman, A. Narcissistic Personality Measures Discriminate Between Young Women With and Without Tattoos. J Forensic Sci & Criminal Inves. 2021; 15(2): 555906 DOI:10.19080/JFSCI.2021.15.555908.
Abstract
In forensic science STR loci are useful tools for reconstructing male lineages, paternity testing, forensic investigations, and population genomics. They cover a wide range of genome-specific geographical distributions due to shortfall of recombination during spermatogenesis. The samples used in this study were received at State Forensic Science Laboratory for routine examination. A combination of autosomal (Power Plex®- Fusion 5Csystem kit) and Y-STR (Power Plex®- Y23 system kit) multiplexing systems was used for the genotyping of the samples as per the manufacturer’s protocol. In electropherogram of male samples, male-derived amelogenin marker was not observed in the results of Power Plex®- Fusion 5C. These samples were further analyzed by Power Plex®- Y23 system kit and a complete set of 23 Y STR markers was obtained. The failure rate of detection of amelogenin marker in Indian population is 0.23%. This clearly indicates the deletion in the short arm of Y chromosome related to amelogenin marker.
Keywords: Amelogenin marker; Forensic genetics; Haplogroups; Deletion
Introduction
DNA analysis based on autosomal as well as sex chromosome STR is routinely used in forensic casework, population studies, medico-legal examinations etc. [1-4]. In humans, sex-determining genes known as amelogenins are present on both the X and the Y chromosomes. A single-copy gene (Amelogenin gene) located on Xp22.1eXp22.3 and Yp11.2 was sequenced by [5]. Length of AMELY and AMELX genes is 3272 bp and 2872 bp respectively. It has been observed that a single reaction amplifies both genes, and therefore, X amelogenin gene should always be present in the reaction which offers a positive control. Amelogenin gene codes for tooth enamel. Enamel formation is not significantly affected by AMELY deletion due to the presence of its counterpart X chromosome, but it helps in forensic interpretation. Difference in size of these chromosomes has been recognized in forensic casework. It offers a remarkable tool for distinguishing between the evidence of victim and offender in sexual assault cases. As a result of AMELY deletion, normal males may be observed as female because of failure in the amplification of the gene. Misinterpretation of the marker may lead to errors in forensic inference. Various studies have been performed showing mutation in AMELY genes. This mutation may lead to serious consequences, if the males are typed as females due to AMELY deletion particularly in cases of rape or human identification [6]. reported 1.85% failure in the amplification of AMELY locus in 270 Indian males through Southern hybridization [5]. Some studies reported that the primer binding site region includes point mutation instead of AMELY mutation [8,9]. Interstitial deletion of AMELY locus at short arm of Y chromosome was also reported [10]. AMELY deletion in various populations has also been reported by several workers viz., in Australian [11], and Italian [12] populations, where Haas-Rocholz and Weiler demonstrated smaller flanking sequences in primer producing PCR fragments using a different method for the determination of gender in null AMELY males [13]. Lack of Y-amelogenin in 6 out of 29,432 (0.02%)Austrian males has also been demonstrated [14]. For the purpose of gender identification, companies are manufacturing various multiplexing systems that incorporate amelogenin marker. Our study using Power Plex®- Fusion 5C system kit inferred a deletion at the short arm of Y chromosome (amelogenin gene) in the male population of Rajasthan. It was determined by using PowerPlex®- Y23 system kit.
Materials and Methods
The samples for the study were taken from the routine casework received at DNA division State Forensic Science Laboratory, Jaipur with prior written informed consent from the candidates as per the declaration of Helsinki and were subjected to DNA isolation by using Prep Filer Express TM kit (Thermo Fisher Scientific, CA, USA-Thermo) on Automate Express system (Thermo) according to the recommended protocol of the manufacturer. Quantification of isolated samples was performed by using Quanti filer Trio kit (Thermo) on Quant Studio 5 system (Thermo). Amplification of 1ng quantified DNA was done using Power Plex®- Fusion 5C system kit for 22 autosomal STRs viz., D3S1358, D2S441, VWA, D21S11, D10S1248, D13S317, PENTA-E, D16S539, THO1, D18S51, D2S1338, TPOX, CSF1PO, PENTA-D, D7S820, D5S818, D8S1179, D12S391, D19S433, FGA, D1S1656andD22S1045. Amplification of Y specific markers was carried out by using Power Plex®- Y23 system kit for 23 Y STRs like Y_GATA_ H4, DYS389I, DYS385a/b, DYS389II, DYS391, DYS481, DYS392, DYS549, DYS533, DYS438, DYS437, DYS570, DYS635, DYS390, DYS439, DYS643, DYS393, DYS458, DYS448, DYS19, DYS456 and DYS576according to the recommended protocol except for half reaction volume. PCR products were subjected to Genetic Analyzer 3500 XL using POPTM-4 (Performance Optimized Polymer), 36cm Capillary array. Each amplified sample was mixed with 9.5 μL Hi-Di Formamide and 0.5 μL ILS 500. Injection of the samples was done at 1.2 kV for 5s. Electrophoresis results were analyzed with GeneMapper ID-X v1.6 software (Thermo).
Result and Discussion
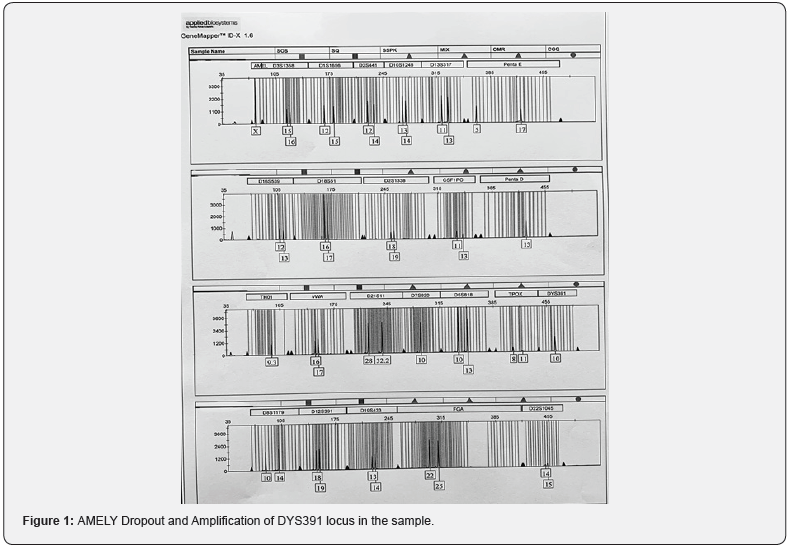
Since X and Y amelogenin markers can be amplified along with autosomal markers in a single reaction, it is used in forensic casework analysis for gender identification. Analysis of the electropherogram of male samples demonstrated a dropout of Y-specific amelogenin (AMEL Y) marker because of amplification of one additional Y specific marker DYS391 in the multiplexing system (Figure 1). This was further reinforced by successful amplification of 23 loci dispersed along both arms of Y-chromosome with the help of PowerPlex®- Y23 system kit. In this manner, our study established the presence of AMEL Ydropouts in the population of Rajasthan which is in concordance with outcomes of similar studies on Indian population [15]. Dropout in AMELY shows pinpoint deletion and the possibility of subsidiary chromosomal aberration needs to be explored. Dropout of AMELY marker encourages the integration of additional Y chromosome specific markers in the multiplexing system. It is more useful in the populations where AMELY dropout frequency is very high. Although frequency of amelogenin dropout is very low (1.85%) in population of Rajasthan, conclusions should not be drawn based on this marker alone. The routinely used Y specific marker DYS389 is most suitable because it explores two loci simultaneously, ranging between 239 to 263 bp and 353-385 bp respectively [16].
Acknowledgment
Authors are grateful to the Director, State Forensic Science Laboratory, Jaipur, Rajasthan for encouraging the study.
Compliance with Ethical Standards
As per the declaration of Helsinki, written informed consent was obtained for the study.
References
- Kumar A, Kumar R, Mohsin U, Sharma S (2019) Genetic Profiling of Short Tandem Repeat (STR) in forensic science to Crackdown complex cases of sexual abuses.
- Kumar A, Kumar R, Kumawat RK, Tilawat A, Shrivastava P, et al. (2020) Genetic variation (population database) at 20 autosomal STR loci in the population of Rajasthan (north-western India). Int J Legal Med 134(5): 1-3.
- Kumar A, Kumar R, Kumawat RK (2020) Genetic portrait study for 23 Y-STR loci in the population of Rajasthan, India. Int J Legal Med 134(5): 1691-1693.
- Kumar A, Kumar R, Kumawat RK, Kumawat G (2021) Maternal allele mutation: Slippage synthesis furnishing evolutionary trend 302016: 90-94.
- Nakahori Y, Hamano K, Iwaya M, Nakagome Y (1991) Sex identification by polymerase chain reaction using X‐Y homologous primer. Am J Med Genet 39(4): 472-473.
- Hart PS, Vlaservich AC, Hart TC, Wright JT (2000) Polymorphism (g2035C> T) in the amelogenin gene. Hum Mutat 15(3): 298.
- Thangaraj K, Reddy AG, Singh L (2002) Is the amelogenin gene reliable for gender identification in forensic casework and prenatal diagnosis. Int J Legal Med 116(2): 121-123.
- Roffey PE, Eckhoff CI, Kuhl JL (2001) A rare mutation in the amelogenin gene and its potential investigative ramifications. J Forensic Sci 45(5): 1016-1019.
- Henke J, Henke L, Chatthopadhyay P, Kayser M, Dulmer M, et al. (2001) Application of Y-chromosomal STR haplotypes to forensic genetics. Croat Med J 42(3): 292-297.
- Lattanzi W, Di Giacomo MC, Lenato GM (2005) A large interstitial deletion encompassing the amelogenin gene on the short arm of the Y chromosome. Hum Genet 116(5): 395-401.
- Mitchell RJ, Kreskas M, Baxter E, Buffalino L, Van Oorschot RAH (2006) An investigation of sequence deletions of amelogenin (AMELY), a Y-chromosome locus commonly used for gender determination. Ann Hum Biol 33(2): 227-240.
- Turrina S, Filippini G, Voglino G, De Leo D (2011) Two additional reports of deletion on the short arm of the Y chromosome. Forensic Sci Int Genet 5(3): 242-246.
- Haas Rochholz H, Weiler G (1997) Additional primer sets for an amelogenin gene PCR-based DNA-sex test. Int J Legal Med 110(6): 312-315.
- Steinlechner M, Berger B, Niederstätter H, Parson W (2002) Rare failures in the amelogenin sex test. Int J Legal Med 116(2): 117-120.
- Kashyap VK, Sahoo S, Sitalaximi T, Trivedi R (2006) Deletions in the Y-derived amelogenin gene fragment in the Indian population. BMC Med Genet 7(1): 37.
- Thangaraj K, Ramana GV, Singh L (1999) Y‐chromosome and mitochondrial DNA polymorphisms in Indian populations. Electrophor An Int J 20(8): 1743-1747.






























