Distinct Pathways of Regulation Seed Dormancy and Germination by Abscisic Acid
Obroucheva NV*, Lityagina SV and Sinkevich IA
Institute of plant physiology of Russian Academy of Sciences, Russia
Submission: December 21, 2018; Published: January 11, 2019
*Corresponding author: Obroucheva NV, Institute of plant physiology of Russian Academy of Sciences, Botanicheskaya str.35, Moscow 127276, Russia
How to cite this article: Obroucheva NV, Lityagina S, Sinkevich I. Distinct Pathways of Regulation Seed Dormancy and Germination by Abscisic Acid. Int J cell Sci & mol biol. 2019; 5(4): 555667. DOI: 10.19080/IJCSMB.2019.05.555667
Abstract
Abscisic Acid (ABA) is the phytohormone inducing seed dormancy, whereas seed germination occurs at low ABA concentration. Two pathways of their regulation are considered. One represents ABA reception and its signal transduction to ABA-responsive genes to induce dormancy genes, a modification of this way leading to germination in the absence of ABA. Another way suggests direct ABA inhibition of plasmamembrane H+-ATPase in dormant seeds that prevents germination and its activation in the absence of ABA, thus promoting seed germination.
Keywords: Seed dormancy; Germination; Transcription of dormancy genes; Activation of plasma membrane H+-ATPase.
Introduction
After the discovery of ABA receptors belonging to PYR/PYL/ RCAR protein family [1,2], and mechanism of their functioning [3], the components of signal system transducing the ABA signal to nuclear ABA-sensitive genes were identified [4,5]. According to the modern point of view, this regulatory pathway is common for the induction of primary seed dormancy and its maintenance. However, dormancy release includes the ABA degradation [6], and non-dormant seeds contain low ABA concentrations, which do not prevent seed germination [7,8]. In the absence of ABA, this signal system is modified to make the germination possible [9,10]. Figure 1 schematically describes such model of ABA regulation established on Arabidopsis seeds. ABA signaling system in seeds consists of three components, namely protein phosphatases, protein kinases and transcription factor ABI5.
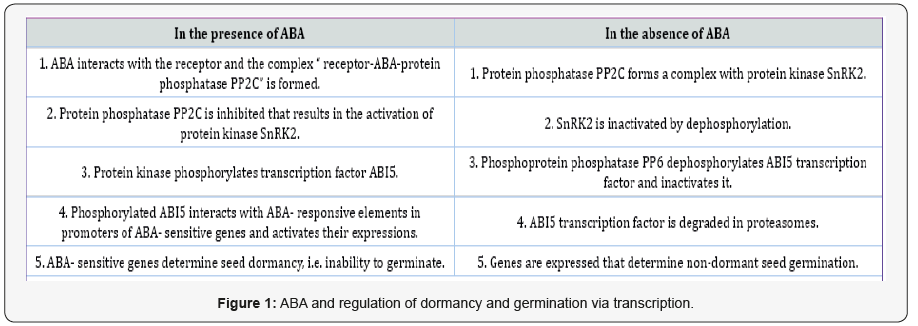
Protein phosphatases PP2C, known also as ABI1 and ABI2 factors, negatively regulate ABA responses. Arabidopsis seeds have six genes of PP2Cs, the expression of which results in the production of these proteins taking part in the formation of the ternary complex with ABA and its receptor and in modulating the phosphorylation activity of protein kinases. Three protein kinases SnRK2 take part in ABA signaling. They mediate phosphorylation and activation of transcription factor ABI5- the step critical in the dormancy and inhibition of germination.
ABI5 protein always has a phosphorylation site to be activated by SnRK2 protein kinases. This ABA-responsive activation is accompanied by its stabilization during dormancy induction and maintenance. The participation of ABI5 permits the expression of ABA-responsive dormancy–specific genes. The degradation of ABI5 is necessary for the initiation of germination. It begins by ABI5 dephosphorylation by another family of serine/threonine protein phosphatases, namely FYPP/PP6 phosphoprotein phosphatases, followed by its breakdown through the 26S proteasome-dependent pathway. ABI5 dephosphorylation is an essential prerequisite for eliminating the inhibitory effect of ABA on germination. The abolishment of ABI5 transcription factor causing subsequent changes in gene expression is a key event for seed germination.
The successful functioning of ABA reception and subsequent transduction of the signal to the corresponding nuclear genes determines the so-called ABA sensitivity; any disturbance of receptor--signaling system, for example in mutants, results in a loss of ABA sensitivity.. The list of genes involved in dormancy regulation was presented by Graeber et al. [11]. In dormant Arabidopsis seeds gene expression is enriched for proteins involved in ABA synthesis, gibberellin catabolism, protein kinases, protein phosphatases, numerous transcription factors, and typical maturation proteins, namely small heat shock proteins and Late Embryogenesis Abundant (LEA) proteins [12,13]. Special attention was paid to DOG1 protein regulating seed dormancy induction and delaying germination by interfering with PP2C protein phosphatase activity [14]. The translation of proteins occurs slowly in dormant seeds.
The set of germination proteins radically differs from the proteins translated during dormancy. In imbibing and germinating seeds, translational machinery operates much more rapidly than in dormant seeds [15]. Protein composition depends first of all on mRNAs stored during maturation [16,17]. At radicle emergence state and after germination, de novo transcription of other genes starts. As a result, germinating seeds contain the complete set of “house-keeping” enzymes providing glycolysis, TCA cycle, gluconeogenesis, glyoxalate cycle, amino-acid metabolism, respiration, pentose phosphate cycle, ribosome functioning and other processes constituting main metabolic and energy utilization systems. They contain also a set of enzymes participating in the mobilization of the reserves (proteins, starch and lipids) as well as aquaporins and gibberellin synthesizing enzymes. Numerous transcription factors are produced while the components of ABA signal system are downregulated. Some maturation proteins still are produced (LEA and heat shock proteins), but stress-proteins and cell cycle proteins appeared. The list of proteins identified in germinating rice embryos is presented in [18].
We have considered the pathway of seed dormancy and germination regulation based either on inhibitory action of ABA on growth initiation resulting in dormancy or on the absence of such ABA action leading to germination commencement (Figure1). This pathway operates through the receptor--signaling–transcription system. However, quite ananother regulatory pathway can function in parallel, namely via inactivation (in the case of dormancy) or activation (in the case of germination) of the enzyme H+-ATPase responsible for H+ ions transfer from cytoplasm through the plasma membrane to cell wall. H+ efflux is accompanied by K+ infux into cytoplasm. The acidified cell walls acquire the ability to activate the enzymes modifying the structure of hemicellulose polymers and of expansin proteins facilitating the displacement of polymer chains relative each other. As a result, activated enzyme increases the extensibility of cell wall, i.e. the prerequisite for growth initiation. The cell extension itself begins under the pressure of water entering cell. In addition, K+ ions delivered to cytoplasm support the osmotic potential and favor water uptake. In the presence of inhibitory ABA concentrations, no activation of plasma membrane H+-ATPase occurs.
Plasma membrane H+-ATPase (proton ATPase) is a transmembrane protein, the most part of the molecule is situated in cytoplasm while five loops are localized outside the membrane [19]. In cytoplasm, both hydrophilic N and C termini as well as three domains are located, namely for binding nucleotides, for ATP hydrolysis and for activity regulation. Two activity states of enzyme are known: an autoinhibited state, when ATP hydrolysis is only loosely coupled to H+ transport, and an upregulated state with tight coupling between ATP hydrolysis and H+ pumping. In autoinhibited state, eight times more ATP is required to pump the same number of H+ ions as in upregulated state. In autoinhibited state, the regulatory C domain is pressed into the intramolecular binding site of protein [20]. Both N and C termini together take part in its detachment, apparently due to a structural rearrangement making the penultimate threonine residue accessible for protein kinase-mediated phosphorylation and subsequent activation of the enzyme [21].
The activation occurs inside the terminal region of regulatory domain at the level of penultimate threonine, the phosphorylation of which creates a binding site for 1-3-3 protein, which stabilizes the upregulated state of the enzyme, i.e. stabilizes its conformation [19]. Fusicoccin, a specific activator of plasma membrane H+-ATPase [22], binds to the enzyme simultaneously with 14-3-3 protein, its molecule being situated within the cavity in the interaction surface between H+-ATPase and 14-3-3 protein to tightly couple both partners [19]. In the upregulated state, the enzyme actively extrudes the H+ ions to cell walls [23]. The activity of plasma membrane H+-ATPase is measured either by rate of ATP hydrolysis, usually by the release of inorganic phosphate, or by H+ ion extrusion, estimated by acidification of ambient solution or by pH shift recorded by indicator color. In the experiments, fusicoccin is applied as a specific activator, whereas vanadate and diethylstilbestrol are applied as the inhibitors of ATP hydrolysis.
The capacity of plasma membrane H+-ATPase to acidify cell walls and promote cell elongation is well-known as an “acid growth” (acid-induced growth) exhibited in coleoptiles and hypocotyls, the growth of which starts after radicle protrusion [24,25]. However, no data on the participation of plasma membrane H+-ATPase in seed germination is available in literature. We have observed the acidification of cell walls in broad bean, wheat and horse chestnut seeds prior to radicle protrusion that is prior to growth beginning in the embryos of germinating seeds [26,27]. Such acidification is similar to the effect of acid buffer, it is stimulated by fusicoccin that confirms the participation of this enzyme. Analogous activation of ATP hydrolysis occurs just before growth initiation, which is blocked by vanadate. These data indicate a direct participation of the enzyme in seed germination. The experiments with seed incubation in the solutions of fusicoccin stimulating radicle emergence or in diethylstilbestrol and vanadate inhibiting it have confirmed the conclusion.
In seeds, proteomic analysis failed to record the presence of plasma membrane H+-ATPase, because the method does not permit to see the proteins of high molecular weight. The mol. wt. of this enzyme is about 100 kDa. The enzyme can be identified by western blotting and application of specific antibodies. It was shown in broad bean embryo axes in the course of imbibition, radicle emergence and growth [27]. Similarly, in non-dormant horse chestnut seeds, the enzyme protein was identified in imbibing, protruding and growing embryo axes. In all samples, 14-3-3 protein was also present [27]. The plasma membrane H+-ATPase protein was also shown in the embryos of dry maize seeds, they exhibit a weak activity after short 5h-imbibition, not accompanied by protein synthesis [28,29]. The presence of the enzyme in dry seeds shows that it was synthesized and accumulated earlier, apparently during seed maturation. In seed embryo axes, the delay of imbibition by PEG-6000, an osmoticum, retards the acidification, which needs about 68- 70% water content for enzyme activation. Therefore, during imbibition the transition of autoinhibited form of H+-ATPase to an active one occurs. We followed this transition in horse chestnut seeds, which experience the state of deep physiological dormancy and then dormancy release to acquire the capacity of rapid germination after four months of cold wet stratification [29]. The long period of dormancy and dormancy release culminated in the enzyme activation and germination indicates rather prolonged time of the transition from autoinhibited form to an active one.
In dormant seeds, the rate of cell wall acidification in embryo axes is rather low at the initial stage of embryo dormancy, but twofold increased after 6th week of stratification up to the level typical of emerged embryos, when seeds experience only coatimposed dormancy [29]. During embryo dormancy, fusicoccin does not exert any stimulating effect either on H+ ion extrusion into cell walls or on the size of embryos themselves. It means that during embryo dormancy plasma membrane H+-ATPase is still autoinhibited. Its activity and responsiveness to fusicoccin significantly rose after embryo dormancy, when water content in embryo increased and cell preparation for germination occurred. Such activation of the enzyme is not related to indolyl-acetic acid as it was shown for coleoptiles and hypocotyls [24,25], i.e. for postgerminative growth, because the dormancy release and germination of horse chestnut seeds do not respond to the treatments by indolyl-acetic acid or gibberellin.
The difference in plasma membrane H+-ATPase behavior over the whole period of dormancy and dormancy release is apparently due to the dynamics of ABA in embryo axis tissues. The content of endogenous ABA manifold increased during the period of embryo dormancy and then declined to very low level by the germination commencement [30]. ABA accumulation correlates in time with the autoinhibited state of the enzyme. The treatment of horse chestnut embryos with 10-7 M ABA did not inhibit the acidification over this time interval, 10-6 M influenced weakly, while 10-5 M ABA actively inhibited the H+ extrusion, apparently because the concentrations of endogenous and exogenous ABA summated. Therefore, the target of ABA inhibitory action on dormant seeds is plasma membrane H+- ATPase resulted in the cessation of H+ ion delivery to cell walls, the prerequisite of cell growth initiation. Decline of endogenous ABA content leads to the activation of the enzyme and commencement of germination. In non-dormant seeds lacking ABA, the transition of the enzyme to active state is related to an increase in water content.
The described pathway of ABA action on seed germination differs from that described above, namely from the receptor – signaling – transcription system, in two aspects: this pathway is shorter (no ABA–receptor activation) and it does not directly depend on the transcription of new mRNA in imbibing seeds, as follows from the experiments with α-amanitin , the inhibitor of the new mRNAs transcription. After radicle protrusion, when the growth of seedling organs occurs, a postgerminative program includes active operation of plasma membrane H+-ATPase [19,31]; the inhibitory action of ABA on their cell elongation is explained by ABA interaction with the receptor, ABA inhibition of protein phosphatase and ABA activation of protein kinase leading to the phosphorylation of an unknown amino acid residue in regulatory domain of the enzyme. This chain of events can result in the inhibition of enzyme, that in its turn causes K+ efflux from cytoplasm and its acidification due to the damage of H+ ion transport by the enzyme [31]. These observations might give insight into the transition from germination program of seeds to postgerminative program of seedling development.
Acknowledgements
In our patient the rare combination of High-risk HPV, Lymphoma and High grade squamous intraepithelial lesion was quite interesting. But the efficient interdisciplinary discussions gave her a wonderful outcome.
References
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324(5930): 1068-1071.
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, et al. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324(5930): 1064-1068.
- Fujii H, Zhu JK (2009) Arabidopsis mutant deficient in 3 abscisic acid- activated protein kinases reveal critical roles in growth, reproduction and stress. Proc Natl Acad Sci USA 106(20): 8380-8385.
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41(5): 697-709.
- Okamoto M, Tatematsu K, Matsui A, Morosawa T, Ishida J, et al. (2010) Genome-wide analysis of endogenous abscisic acid-mediated transcription of dry and imbibed seeds of Arabidopsis using tilling arrays. Plant J 62(1): 39-51.
- Preston J, Tatematsu K, Kanno Y, Hobo T, Kimura M, et al. (2009) Temporal expression patterns of hormone metabolism genes during imbibition of Arabidopsis thaliana seeds: a comparative study on dormant and non-dormant accessions. Plant Cell Physiol. 50(10): 1786-1800.
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, et al. (2009) Three Arabidopsis SnRK2 protein kinases involved in ABA signaling and essential for the control of seed development and dormancy. Plant Cell Physiol. 50(7): 1345-1363.
- Dai M, Xue G, Mccray T, Margavage K, Chen F, et al. (2013) The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 25(2): 517-534.
- Dai M, Xue G, Mccray T, Margavage K, Chen F, et al. (2013) The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 25(2): 517-534.
- Yoshida T, Mogami J, Yamaguchi-Shinozaki K, Yamaguchi Shinozaki K (2015) Omics approaches toward defining the comprehensive abscisic acid signaling network in plants. Plant Cell Physiol 56(6): 1043-1052.
- Graeber K, Nakabayashi K, Miatton E, Leubner Metzger G, Soppe WJJ, et al. (2012) Molecular mechanisms of seed dormancy. Plant Cell Environ 35(10): 1769-1786.
- Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Ann Rev Plant Biol 59: 387-415.
- Chibani K, Ali Rachedi S, Job C, Jullien M, Grappin P (2006) Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiol 142(4): 1493- 1510.
- Nishimura N, Tsuchiya W, Moresco JJ, Hayashi Y, Schroeder JT, et al. (2018) Control of seed dormancy and germination by DOG1--AHG1 PP2C phosphatase complex via binding to heme. Nat Commun 9(1): 2132-2141.
- Galland M, Huguet R, Arc E, Gueff G, Job D, et al. (2014) Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Mol Cell Proteomics 13(1): 252-268.
- Kimura M, Nambara E (2010) Stored and neosynthesized mRNA in Arabidopsis seeds: effects of cycloheximide and controlled deterioration treatment on the resumption of transcription during imbibition. Plant Mol Biol 73(1-2): 119-129.
- Rajjou L, Gallardo K, Debaujon I, Vandekerckhove J, Job C, et al. (2004) The effect of α-amanitin on the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134(4): 1598-1613.
- Han C, He D, Li M, Yang P (2014) In-depth proteomic analysis of rice embryo results its important roles in seed germination. Plant Cell Physiol 55(10): 1826-1847.
- Falhof J, Redersen JT, Fuglsang AT, Palmgren M (2016) Plasma membrane H+-ATPase regulation in the center of plant physiology. Mol Plant 9(3): 323-337.
- Portillo F (2000) Regulation of plasma membrane H+-ATPase in fungi and plants. Biochem Biophys Acta 1469(1): 31-42.
- Ekberg K, Palmgren MG, Veierskov B, Buch Pedersen J (2010) A novel mechanism of P-type ATPase autoinhibition involving both termini of the protein. J Biol Chem 285(10): 7344-7350.
- Camoni L, Visconti S, Aducci P (2013) The phytotoxin fusicoccin, a selective stabilizer of 14-3-3 interactions? Int. Union Biochem. Mol Biol 65(6): 513-517.
- Buch Pedersen MJ, Pedersen BP, Veierskov B, Nissen P, Palmgren MG (2009) Protons and how they are transported by proton pumps. Eur J Physiol 457(3): 573-579.
- Takahashi K, Hayashi K, Kinoshita T (2012) Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol 159(2): 632-641.
- Hayashi Y, Takahashi K, Innoue S, Kinoshita T (2014) Abscisic acid suppresses hypocotyl elongation by dephosphorylating plasma membrane H+-ATPase in Arabidopsis thaliana. Plant Cell Physiol 55(4): 845-853.
- Obroucheva NV, Sinkevich IA, Lityagina SV (2013) Activation of “acid growth” in germinating horse chestnut seeds. Russ J Plant Physiol 106(3): 41-46.
- Obroucheva NV (2017) Participation of plasma membrane H+-ATPase in seed germination. Int J Cell Sci Mol Biol 2(3): 1-4.
- Sanchez Nieto S, Gomez Puyou MT, Rodriguez Sotres R, Garballo A, Gavilanes Ruiz M (1998) Comparison of plasma membrane H+-ATPase activity in vesicles obtained from dry and hydrated maize embryos. Biochem Biophys Acta 1414 (1-2):175-187.
- Sanches Nieto S, Enriquez Arredondo C, Guzman Chavez F, Hernandez Munoz R, Ramirez J et al. (2011) Kinetics of the H+-ATPase from dry and 5-hours-imbibed maize embryos in its native, solubilized and reconstituted forms. Mol Plant 4(3): 505-515.
- Obroucheva NV, Antipova OV (2000) The distinct controlling of dormancy release and germination commencement in seeds. In: Dormancy in plants: from whole plant behaviour to cellular control, ed. by J.-D. Viemont, J.Crabbe, Wallingford, CABI publ, pp. 35-46.
- Planes M, Ninoles R, Rubio L, Rissoli G, Rueso E, et al. (2015) A mechanism of growth inhibition by abscisic acid in germinating seeds of Arabidopsis thaliana based on inhibition of plasma membrane H+-ATPase and decreased cytosolic pH, K+ and anions. J Exp Bot 66(3): 813-825.






























