Assessment of Genetics Mutation of SMN1, SMA And SMN2 Genes In Spinal Atrophy-Muscle
Asadi S*
Department of Molecular Geneticist, Islamic Azad University, Iran
Submission: September 09, 2017; Published: October 03, 2017
*Corresponding author: Asadi S, Department of Molecular Geneticist, Director of the Division of Medical Genetics and Molecular Research, IRAN- TABRIZ, Islamic Azad University, Iran, Tel: +989379923364; Email: shahin.asadi1985@gmail.com
How to cite this article: Asadi S. Assessment of Genetics Mutation of SMN1, SMA And SMN2 Genes In Spinal Atrophy-Muscle. Int J cell Sci & mol biol. 2017; 3(3) : 555615. DOI: 10.19080/IJCSMB.2017.03.555615.
Abstract
Muscle atrophy syndrome - Spinal (SMA) is one of the common diseases of muscle - nerve, with progressive paralysis is due to the alpha motor neuron in the spinal cord becomes waste. SMN1 and SMN2 gene expression in SMA by only a single nucleotide in exon 7 are different. Homozygous deletion of exon 7 in the SMN1 gene is the most common mutation observed. Compound heterozygosity small proportion of patients with a point mutation in one allele and the other allele are removed. In other cases the disease does not appear to be the result of a change in SMN1. In spinal atrophy-muscle, SMN2 unable to compensate for the shortage caused by the deletion of exon 7. The aim of the present study was to estimate the prevalence of common deletion of exon 7 in the SMN1 gene families in Tabriz, in order to determine the status of the carrier and prenatal diagnosis. 119 families with a history of child with SMA and determined that the deletion of exon 7 in the child with the parents were carriers and Also well as 42 cases of prenatal diagnosis for couples fetus was a carrier. Of the families surveyed, 127 families with a history of SMA1, 24 families with a history of type II and 11 families with a history of type III disease who have had children. In families with a history of a patient, 89 families had both parents carry the deletion of exon 7 and in 26 families, one of them was carrying. The frequencies in families with a history of type II SMA 16 and 1, respectively, and in families with a history of type III SMA was 2,3 respectively. Investigation of fetal samples showed that 11 of 63 samples (5.17%) were diagnosed. Except for two, all patients were homozygote for deletion of exon 7 (7.96% of patients). The study showed that molecular analysis of the two alleles of exon 7 SMN1 appropriate method for the removal and reliable diagnosis of patients to determine the carrier and prenatal diagnosis in families with patients with SMA is.
Keywords: SMN1; SMN2; SMA; Exon 7; PCR
Introduction
SMA Spinal muscular atrophy is a group of genetic diseases that are associated with the analysis, causing muscle weakness and spinal cord motor neurons in most cases leads to death of the infected person [1]. This disease is the most common autosomal recessive disorder after cystic fibrosis The prevalence is considered to have a baby every 10,000 births and carrier frequency of one in four to one in fifty people is different [2]. SMA anterior horn cells of the spinal cord will be determined by analyzing [3]. In this disease, the nerve cells that control muscle movements proximal voluntary, mobility destroyed [4]. That may result from intrauterine period or at any time after it is created and garlic have fast or slow [5]. The earlier the disease starts to progress faster [6]. Infants infected at birth or in the first few months of life are poor usually suffer from paralysis and crippled limbs throat area, respiratory failure and Death in the first few years of life are the early and premature death, disease (werding hoffman) is said to be [7]. Mild form of the disease (Kogelberg Volandri syndrome) in late childhood or adolescence with muscle weakness and upper legs started slowly progresses over several decades [8]. SMA types that usually occurs between the ages of course is unpredictable [9].
Proximal spinal muscular atrophy (SMA) is an autosomal recessive disease caused by a genetic defect in the SMN1 gene, which encodes SMN, a protein widely expressed in all eukaryotic cells. SMN is apparently selectively necessary for survival of motor neurons, as diminished abundance of the protein results in loss of function of neuronal cells in the anterior horn of the spinal cord and subsequent system-wide muscle wasting (atrophy) [10-12].
Spinal muscular atrophy manifests in various degrees of severity, which all have in common progressive muscle wasting and mobility impairment. Proximal muscles and lung muscles are affected first. Other body systems may be affected as well, particularly in early-onset forms. SMA is the most common genetic cause of infant death.
The term spinal muscular atrophy is used as both a specific term for the genetic disorder caused by deficient SMN, and a general label for a larger number of rare disorders having in common a genetic cause and slow progression of weakness without sensory impairment caused by disease of motor neurons in the spinal cord and brainstem-see spinal muscular atrophies for a comparison chart [13-16].
Materials and Methods
In this study, 10 patients with spinal atrophy-muscle disease and 20 persons control group were studied. Peripheral blood samples from patients and parents with written permission control were prepared. After separation of serum, using Real Time-PCR technique of tRNA molecules was collected. To isolate Neuroglial cells erythrocytes were precipitated from hydroxyethyl starch (HES) was used. At this stage, HES solution in ratio of 1 to 5with the peripheral blood of patients and controls were mixed. After 60 minutes of incubation at room temperature, the supernatant was removed and centrifuged for 14min at 400 Gera. The cells sediment with PBS (phosphate buffered saline), pipetazh and slowly soluble carbohydrate ratio of 1to2 on ficole (Ficol) was poured in the 480G was centrifuged for 34 minutes. Mono nuclear Neuroglial cells also are included, has a lower density than ficole and soon which they are based. The remaining erythrocytes has a molecular weight greater than ficole and deposited in test tubes.
The supernatant, which contained the mono nuclear cells was removed, and the 400 Gera was centrifuged for 12 minutes. Finally, the sediment cell, the antibody and Neuroglial cells was added after 34 minutes incubation at 5 °C, the cell mixture was passed from pillar LSMACS. Then the cells were washed with PBS and attached to the column LSMACSS pam Stem cell culture medium containing the transcription genes SMN1, SMN2, SMA, and were kept.
To determine the purity of Neuroglial cells are extracted, flow cytometry was used. For this purpose, approximately 4- 5x103 Neuroglial cells were transfer red to1.5ml Eppendorf tube and then were centrifuged at 2000rpm for 7 minutes at time. Remove the supernatant culture medium and there maining sediment, 100μl of PBS buffer was added. After adding 5- 10μl PE monoclonal anti body to the cell suspension for 60min at 4 °C, incubated and read immediately by flow cytometry. For example, rather than control anti body Neuroglial cells PE, IgG1 negative control solution was used.
Total mRNA extraction procedure includes:
1) 1ml solution spilled Qiazolon cells, and slowly and carefully mixed and incubated at room temperature for 5 minutes. Then 200μl chloroform solution to target mix, and then transfer the micro tubes was added, and the shaker well was mixed for 15 seconds. The present mix for 4 minutes at room temperature and then incubated for 20min at 4 °C on was centrifuged at 13200 rpm era. Remove the upper phase product was transfer reductase new micro tube and to the one times the volume of cold ethanol was added. The resulting mixture for 24 hours at -20 °C was incubated.
2) Then for 45min at 4 °C on was centrifuged at 12000rpm era. Remove the supernatant and the white precipitate, 1ml of cold 75% ethanol was added to separate the sediment from micro tubes were vortex well. The resulting mixture for 20min at 4 °C on by the time we were centrifuged 12000rpm. Ethanol and the sediment was removed and placed at room temperature until completely dry deposition. The precipitate was dissolved in 20μl sterile water and at a later stage, the concentration of extracted mRNA was determined.
Results
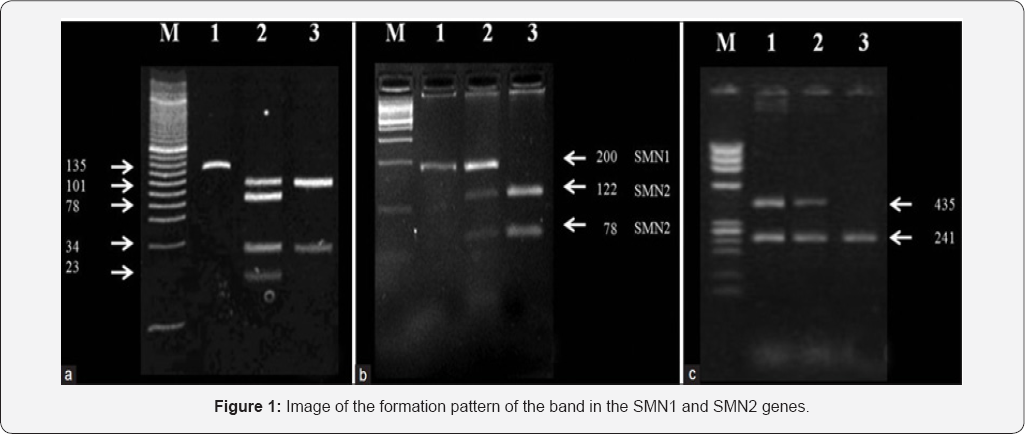
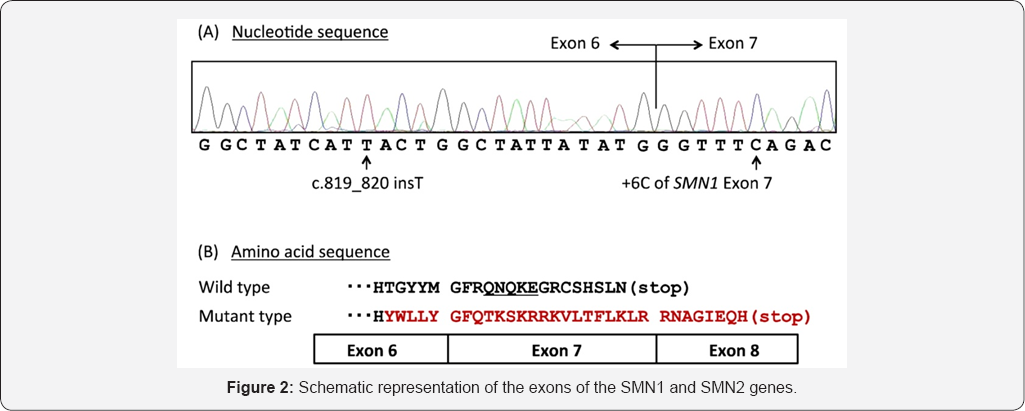
Discussion and conclusion
According to the results of sequencing the genome of patients with spinal atrophy-muscle disease, and the genetic mutations SMN1, SMN2, SMA genes found that about 100% of patients with spinal atrophy-muscle disease, they have these genetic mutations. Patients with spinal atrophy-muscle disease, unusual and frightening images in the process of spinal atrophy- muscle disease, experience. Lot epigenetic factors involved in spinal atrophy-muscle disease. But the most prominent factor to induce spinal atrophy-muscle disease, mutations is SMN1, SMN2, SMA genes. These genes can induce the birth and can also be induced in the adulthood.
References
- Brzustowicz LM, Lehner T, Castilla LH, Penchaszadeh GK, Wilhelmsen KC, et al. (1990) Genetic mapping of chronic childhood-onset spinal muscular atrophy to chromosome 5q11.2-13.3”. Nature 344 (6266): 540-541.
- J^drzejowska M, Milewski M, Zimowski J, Borkowska J, Kostera- Pruszczyk A, et al. (2009) Phenotype modifiers of spinal muscular atrophy: The number of SMN2 gene copies, deletion in the NAIP gene and probably gender influence the course of the disease. Acta Biochimica Polonica 56(1): 103-108.
- Little SE, Janakiraman V, Kaimal A, Musci T, Ecker J, et al. (2010) The cost-effectiveness of prenatal screening for spinal muscular atrophy Am J Obstet Gynecol 202 (3): 253.2e1-e7.
- Rutkove SB, Shefner JM, Gregas M, Butler H, Caracciolo J, et al. (2010) Characterizing spinal muscular atrophy with electrical impedance myography. Muscle Nerve 42(6): 915-921.
- Main M, Kairon H, Mercuri E, Muntoni F (2003) The Hammersmith Functional Motor Scale for Children with Spinal Muscular Atrophy: A Scale to Test Ability and Monitor Progress in Children with Limited Ambulation. Eur J Paediatr Neurol 7(4): 155-159.
- O'Hagen JM, Glanzman AM, McDermott MP, Ryan PA, Flickinger J, et al. (2007) An expanded version of the Hammersmith Functional Motor Scale for SMA II and III patients. Neuromuscul Disord 17(9-10): 693697.
- Glanzman AM, O'Hagen JM, McDermott MP, Martens WB, Flickinger J, et al. (2011) Validation of the Expanded Hammersmith Functional Motor Scale in Spinal Muscular Atrophy Type II and III. J Child Neurol 26(12): 1499-1507.
- Bach JR, Saltstein K, Sinquee D, Weaver B, Komaroff E (2007) LongTerm Survival in Werdnig-Hoffmann Disease. Am J Phys Med Rehabil 86(5): 339.
- Messina S, Pane M, De Rose P, Vasta I, Sorleti D, et al. (2008) Feeding problems and malnutrition in spinal muscular atrophy type II. Neuromuscular Disord 18(5): 389-393.
- Chen YS, Shih HH, Chen TH, Kuo CH, Jong YJ (2011) Prevalence and Risk Factors for Feeding and Swallowing Difficulties in Spinal Muscular Atrophy Types II and III. J Pediatr 160(3): 447-451.
- Tein I, Sloane AE, Donner EJ, Lehotay DC, Millington DS, et al. (1995) Fatty acid oxidation abnormalities in childhood-onset spinal muscular atrophy: Primary or secondary defect(s)? Pediatric neurology 12(1): 21-30.
- Rudnik-Schoneborn S, Heller R, Berg C, Betzler C, Grimm T, et al. (2008) Congenital heart disease is a feature of severe infantile spinal muscular atrophy. Journal of Medical Genetics 45(10): 635-638.
- Laufersweiler-Plass C, Rudnik-Schöneborn S, Zerres K, Backes M, Lehmkuhl G, et al. (2002) Behavioural problems in children and adolescents with spinal muscular atrophy and their siblings. Dev Med Child Neurol 45(1): 44-49.
- Tiziano FD, Lomastro R, Pinto AM, Messina S, d'Amico A, et al. (2010) Salbutamol increases survival motor neuron (SMN) transcript levels in leucocytes of spinal muscular atrophy (SMA) patients: Relevance for clinical trial design. J Med Gene 47(12): 856-858.
- Pane M, Staccioli S, Messina S, d'Amico A, Pelliccioni M, et al. (2008) Daily salbutamol in young patients with SMA type II. Neuromuscul Disord 18(7): 536-540.
- Morandi L (2013) P.6.4 Salbutamol tolerability and efficacy in adult type III SMA patients: Results of a multicentric, molecular and clinical, double-blind, placebo-controlled study. Neuromuscular Disorders 23(9-10): 771.






























